鸡尾酒化学标记用于骨髓来源树突状细胞的深度表面体分析
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
细胞表面蛋白(CSPs)是细胞类型和状态的重要标识符,特别是在树突状细胞(dc)中。目前用于分析csp的蛋白质组学方法受到其疏水性和低丰度的限制,这通常需要遗传或细胞表面工程,并且表现出偏见和不足的标记效率。在这里,我们报道了[Ru(bpy)3]Cl2 (Ru)通过简单的“混合和变亮”方法在细胞表面进行有效的生物素化。利用生物素-苯酚和生物素-肼的探针混合物来改善底物覆盖,利用Ru的多用途光氧化还原途径。“鸡尾酒”标记策略在HeLa细胞上可重复识别多达733种质膜蛋白,并进一步应用于绘制原代骨髓来源树突状细胞分化过程中表面体的动态变化,这证明了一种用户友好且深度表面体覆盖的工具,可用于分析原代细胞的动态变化,对细胞身份,功能状态和新的药物靶点具有潜在的意义。本文章由计算机程序翻译,如有差异,请以英文原文为准。
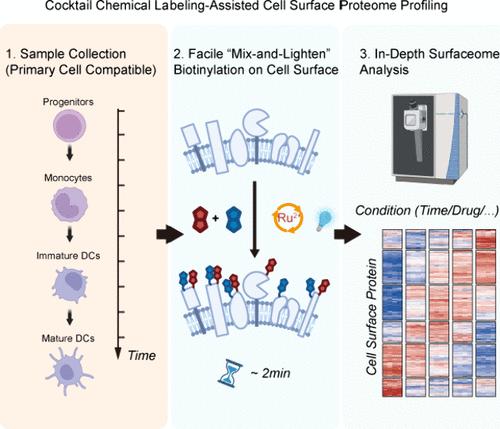
Cocktail Chemical Labeling for In-Depth Surfaceome Profiling of Bone-Marrow-Derived Dendritic Cells
Cell surface proteins (CSPs) are crucial identifiers for cell types and states, especially in dendritic cells (DCs). Current proteomic methods for profiling CSPs are limited by their hydrophobic nature and low abundance, which often require genetic or cell surface engineering and exhibit biased and insufficient labeling efficiency. Herein, we report [Ru(bpy)3]Cl2 (Ru) for effective biotinylation on the cell surfaceome via a simple “mix and lighten” method. The versatile photoredox pathways of Ru are leveraged using a probe cocktail of biotin-phenol and biotin-hydrazide for improved substrate coverage. The “cocktail” labeling strategy results in reproducible identification of up to 733 plasma membrane proteins on HeLa cells and is further applied to map dynamic changes in the surfaceome during the differentiation of primary bone-marrow-derived dendritic cells, which demonstrates a user-friendly and deep-surfaceome-coverage tool for profiling dynamic changes in primary cells, with potential implications for cell identities, functional states, and novel drug targets.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


