利用与人工智能集成的MALDI-TOF质谱直接检测和分化哈维弧菌分支,实现有效的疫情管理
IF 9.8
1区 农林科学
Q1 CHEMISTRY, APPLIED
引用次数: 0
摘要
准确鉴定哈威氏弧菌分支对海产品安全和水产养殖疾病的控制具有重要意义。然而,现有方法的分类性能有限。本研究提出了一种人工智能辅助基质辅助激光解吸电离飞行时间质谱(MALDI-TOF MS)方法,利用板上蛋白质提取进行直接菌落分析。在评估的6种算法中,随机森林模型获得了很好的准确率,显著优于商业数据库,后者的准确率仅为79.57 %。主成分分析表明,当使用模型选择的前10个 %的特征时,物种分离得到了改善。接收机工作特性和查准率曲线进一步证实了模型的鲁棒性和泛化性。7194.7、3609.8和9179.6 m/z处的关键质量峰被确定为主要的区别特征。该方法提供了一种快速、独立于数据库的方法来分类哈氏弧菌分支,增强了MALDI-TOF质谱在食品安全中的应用,并支持其在日常质量控制中的应用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
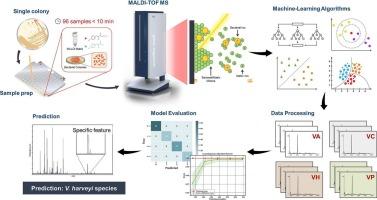

Direct detection and differentiation of the Vibrio harveyi clade using MALDI-TOF MS integrated with artificial intelligence for effective outbreak management
Accurate identification of Vibrio harveyi clade species is critical for seafood safety and the control of aquaculture diseases. However, existing methods demonstrate limited classification performance. This study presents an artificial intelligence-assisted matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI–TOF MS) approach using on-plate protein extraction for direct colony analysis. Among six evaluated algorithms, the random forest model achieved perfect accuracy, significantly outperforming commercial databases, which showed only 79.57 % accuracy. Principal component analysis revealed improved species separation when using the top 10 % of features selected by the model. Receiver operating characteristic and precision-recall curves further confirmed the model's robustness and generalizability. Key mass peaks at 7194.7, 3609.8, and 9179.6 m/z were identified as major discriminatory features. This method offers a rapid, database-independent solution for classifying the V. harveyi clade species, enhancing MALDI–TOF MS utility in food safety and supporting its use in routine quality control.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Food Chemistry
工程技术-食品科技
CiteScore
16.30
自引率
10.20%
发文量
3130
审稿时长
122 days
期刊介绍:
Food Chemistry publishes original research papers dealing with the advancement of the chemistry and biochemistry of foods or the analytical methods/ approach used. All papers should focus on the novelty of the research carried out.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


