Orbitrap星质谱计非靶向代谢组学数据的增强结构引导分子网络注释方法
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
快速、高效和准确地标注复杂样品中的化合物仍然是代谢组学的一个重大挑战。最近开发的Orbitrap星质谱计(MS)集成了传统的四极轨Orbitrap和新型的星质分析仪,提供快速的MS/MS扫描速度和高灵敏度。然而,现有的代谢组学注释方法并没有充分利用Astral MS的先进功能。本研究开发了一种专门为Orbitrap Astral质谱计(MS)量身定制的增强型结构引导分子网络(E-SGMN)方法。与以往的网络标注方法不同,E-SGMN既提取了代谢组数据库中已检测到的代谢物,也提取了Astral可能检测到的代谢物,通过结构相似性实现了更高效、准确的网络构建。E-SGMN通过准确地提高网络规模来扩大标注覆盖范围,同时最大限度地减少不相关化合物的包含,实现标注规模和准确性之间的平衡。验证结果显示,Astral-E-SGMN对加标血浆的标注覆盖率和准确率分别达到76.84%和78.08%,显著优于E-SGMN-Q Exactive HF (E-SGMN-QE HF)。值得注意的是,来自NIST SRM 1950人血浆的5440个代谢物特征被Astral-E-SGMN注释,比QE HF-SGMN增加了3.6倍。对6种典型生物样本的对比分析表明,E-SGMN- astral的代谢物注释能力是传统注释方法的3.7-44.2倍,突出了E-SGMN的代谢物注释覆盖面更广。该方法不仅提高了标注的覆盖率,而且为理解复杂的生物系统提供了一种变革性的工具,在生命科学和临床医学领域具有重要的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
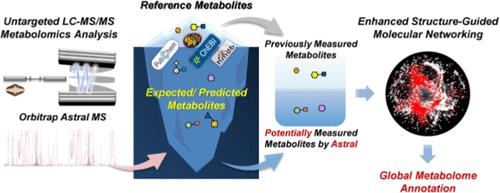
Enhanced Structure-guided Molecular Networking Annotation Method for Untargeted Metabolomics Data from Orbitrap Astral Mass Spectrometer
The rapid, efficient, and accurate annotation of compounds in complex samples remains a significant challenge in metabolomics. The recently developed Orbitrap Astral mass spectrometer (MS) integrates a traditional quadrupole Orbitrap with a novel Astral mass analyzer, providing fast MS/MS scanning speed and high sensitivity. However, existing metabolomics annotation methods have not fully exploited the advanced capabilities of Astral MS. In this study, an enhanced structure-guided molecular networking (E-SGMN) method was developed, which is specifically tailored for the Orbitrap Astral mass spectrometer (MS). Unlike previous network annotation methods, E-SGMN extracted both previously detected metabolites and those potentially detected by Astral from the metabolome database, enabling more efficient and accurate network construction through structural similarity. E-SGMN expands annotation coverage by accurately improving network size, while minimizing the inclusion of irrelevant compounds, achieving a balance between annotation scale and accuracy. Validation results revealed that Astral-E-SGMN achieved an annotation coverage and accuracy of 76.84% and 78.08%, respectively, for a spiked plasma, significantly outperforming E-SGMN-Q Exactive HF (E-SGMN-QE HF). Notably, 5440 metabolite features from NIST SRM 1950 human plasma were annotated by Astral-E-SGMN, a 3.6-fold increase over QE HF-SGMN. Comparative analyses for six types of typical biological samples demonstrate that E-SGMN-Astral enhanced metabolite annotations by 3.7–44.2 times compared to conventional annotation methods, highlighting E-SGMN’s wider metabolite annotation coverage. This method not only enhances annotation coverage, but also provides a transformative tool for understanding complex biological systems, holding significant potential for life science and clinical medicine.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


