细胞外小泡MALDI-TOF MS指纹图谱快速鉴定食管鳞状细胞癌生物标志物
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
食管癌是一项重大的全球健康挑战,由于缺乏快速、敏感的诊断工具和特异性生物标志物,食管癌的发病率和死亡率都很高。癌细胞来源的细胞外囊泡(ev)携带独特的蛋白质和核酸,使其成为癌症生物标志物的宝贵来源。我们报道了一种结合超快速外泌体分离系统(EXODUS)和基质辅助激光脱附电离飞行时间质谱(MALDI-TOF MS)的综合方法,用于检测ev和鉴定诊断和监测食管鳞状细胞癌(ESCC)的蛋白质生物标志物。从不同分化程度的ESCC细胞20 mL培养基上清液中获得的ev作为分析模型。我们使用EXODUS快速分离电动汽车。然后,我们使用MALDI-TOF质谱分析完整的EV,在几分钟内提供细胞系特异性EV指纹。这些蛋白指纹图谱可以区分ESCC和正常对照细胞,并可以根据细胞分化程度对ESCC进行分类。我们探索了ESCC诊断(5555 m/z, 8603 m/z等)和监测(2268 m/z等)的关键EV生物标志物峰。潜在的EV生物标志物候选物,包括YBX1, DIRAS2, HIST1H2AH和MYBBP1A,通过串联质量标签(TMT)蛋白质组学鉴定。我们通过与TMT蛋白质组学的相关性初步确定了EV标记峰的蛋白质身份。将该方法应用于血浆源性EVs,有望实现ESCC的快速、微创诊断和监测。本文章由计算机程序翻译,如有差异,请以英文原文为准。
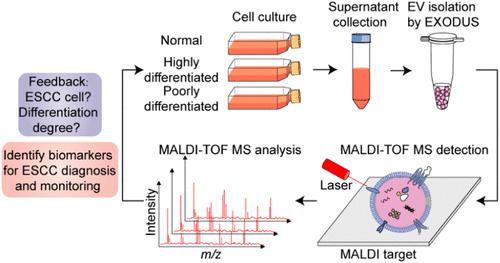
Rapid Identification of Esophageal Squamous Cell Carcinoma Biomarkers by MALDI-TOF MS Fingerprinting of Extracellular Vesicles
Esophageal cancer is a major global health challenge, with high incidence and mortality due to the lack of rapid and sensitive diagnostic tools and specific biomarkers. Cancer-cell-derived extracellular vesicles (EVs) carry unique proteins and nucleic acids, making them valuable sources of cancer biomarkers. We report an integrated method that combines an ultrafast exosome isolation system (EXODUS) with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) to detect EVs and identify protein biomarkers for diagnosing and monitoring esophageal squamous cell carcinoma (ESCC). EVs derived from 20 mL culture medium supernatant of ESCC cells with varying degrees of differentiation serve as analysis models. We use EXODUS to isolate EVs rapidly. We then analyze the intact EVs using MALDI-TOF MS, which provides cell line-specific EV fingerprints in minutes. These protein fingerprints allow the discrimination of ESCC from normal control cells and enable the classification of ESCC based on the degree of cell differentiation. We explore critical EV biomarker peaks for ESCC diagnosis (5555 m/z, 8603 m/z, etc.) and monitoring (2268 m/z, etc.). Potential EV biomarker candidates, including YBX1, DIRAS2, HIST1H2AH, and MYBBP1A, are identified through tandem mass tag (TMT) proteomics. We tentatively assign the protein identities of EV marker peaks by correlation with the TMT proteomics. Applying this method to plasma-derived EVs shows promise for rapid, minimally invasive diagnosis and monitoring of ESCC.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


