基于结构设计同时提高铜绿假单胞菌氨基肽酶的催化活性和热稳定性
IF 6.2
1区 农林科学
Q1 AGRICULTURE, MULTIDISCIPLINARY
引用次数: 0
摘要
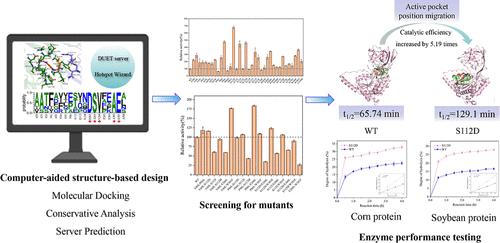
氨基肽酶是食品和制药工业中至关重要的水解酶。本研究通过多方面的计算设计策略来解决提高铜绿假单胞菌氨基肽酶(PaAps)催化性能的需要。我们通过引入单位点突变和组合突变建立突变文库,确定了最佳突变体S112D,与野生型相比,该突变体的催化活性提高了5.19倍,热稳定性提高了近一倍。S112D的动力学参数kcat、kcat/Km和Vmax分别是野生型的4.36倍、6.52倍和4.36倍。分子动力学(MD)模拟显示,S112D突变体诱导了全局构象变化,导致更开放的活性口袋,促进了与底物的更好结合,从而提高了构象稳定性。此外,突变体S112D表现出更近的亲核攻击距离和更强的氢键相互作用,进一步提高了催化效率。值得注意的是,突变体S112D和野生型都表现出对玉米和大豆蛋白的水解活性。S112D对玉米蛋白的水解率约为PaAps的1.92倍,对大豆蛋白的水解率约为1.84倍。这些发现为开发更有效的酶修饰策略提供了有价值的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Simultaneously Enhancing the Catalytic Activity and Thermostability of Pseudomonas aeruginosa Aminopeptidase via Structure-based Design
Aminopeptidases are crucial hydrolases in the food and pharmaceutical industries. This study addresses the need to enhance the catalytic performance of Pseudomonas aeruginosa aminopeptidase (PaAps) through a multifaceted computational design strategy. We introduced single-site mutations followed by combinatorial mutations to develop a mutant library, identifying the optimal mutant S112D, which demonstrated a 5.19-fold increase in catalytic activity and nearly doubled the thermostability compared to the wild type. The kinetic parameters (kcat, kcat/Km, and Vmax) of S112D were found to be 4.36, 6.52, and 4.36 times greater than those of the wild type, respectively. Molecular dynamics (MD) simulations revealed that the S112D mutant induced global conformational changes, resulting in a more open active pocket that facilitated better binding with the substrate, thereby improving conformational stability. Additionally, the S112D mutant exhibited a closer nucleophilic attack distance and stronger hydrogen bonding interactions, further boosting catalytic efficiency. Remarkably, mutant S112D, as well as the wild type, showed hydrolytic activity on both corn and soybean proteins. The hydrolysis rate of corn protein by S112D was approximately 1.92 times that of PaAps, and for soybean protein, it is roughly 1.84 times. These findings offered valuable insights for developing more efficient enzyme modification strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
9.90
自引率
8.20%
发文量
1375
审稿时长
2.3 months
期刊介绍:
The Journal of Agricultural and Food Chemistry publishes high-quality, cutting edge original research representing complete studies and research advances dealing with the chemistry and biochemistry of agriculture and food. The Journal also encourages papers with chemistry and/or biochemistry as a major component combined with biological/sensory/nutritional/toxicological evaluation related to agriculture and/or food.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


