α-琼脂酶CmAga催化机制与多结构域协同作用的结构分析
IF 6.2
1区 农林科学
Q1 AGRICULTURE, MULTIDISCIPLINARY
引用次数: 0
摘要
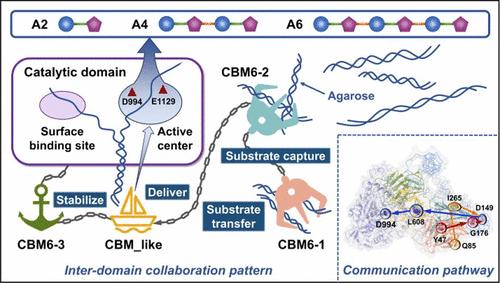
α-琼脂酶是一种糖苷水解酶,它可以裂解琼脂糖中的α-1,3-糖苷键,生成具有生物活性的琼脂低聚糖。尽管α-琼脂酶具有巨大的工业潜力,但由于其结构复杂而灵活,其结构和功能机制尚不清楚。本研究采用Cryo-EM和AlphaFold2相结合的方法,研究了Catenovulum marium STB14 α-琼脂酶CmAga的结构催化机制。D994和E1129被鉴定为催化残基,E1129选择性识别α-1,3-糖苷键。Y858、W1201、Y1164和W1166在−3 ~ +3亚位促进优先底物结合。分子动力学模拟和神经关联推理模型揭示了催化结构域(CD)和四个碳水化合物结合模块(CBMs)的协同机制,CBM6-1和CBM6-2捕获底物,CBM_like将底物转移到CD上,CBM6-3稳定活性位点。D149和L608是域间通信通路的关键节点。这些见解为具有多个CBMs的碳水化合物活性酶(CAZymes)的机理研究和合理工程设计提供了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structure-Informed Insights into Catalytic Mechanism and Multidomain Collaboration in α-Agarase CmAga
α-Agarases are glycoside hydrolases that cleave α-1,3-glycosidic bonds in agarose to produce bioactive agarooligosaccharides. Despite their great industrial potential, the structures and functional mechanisms of α-agarases remain unclear due to their complex and flexible architecture. Here, we investigated the structure-based catalytic mechanism of α-agarase CmAga from Catenovulum maritimum STB14 by integrated Cryo-EM and AlphaFold2. D994 and E1129 were identified as catalytic residues, with E1129 selectively recognizing α-1,3-glycosidic bonds. Y858, W1201, Y1164, and W1166 facilitate preferential substrate binding at the −3 ∼ +3 subsites. Molecular dynamics simulations and neural relational inference modeling revealed a cooperative mechanism involving the catalytic domain (CD) and four carbohydrate-binding modules (CBMs), with CBM6–1 and CBM6–2 capturing substrates, CBM_like transferring them to the CD, and CBM6–3 stabilizing the active site. D149 and L608 served as pivotal nodes within the interdomain communication pathways. These insights provide a foundation for mechanistic investigations and rational engineering of carbohydrate-active enzymes (CAZymes) with multiple CBMs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
9.90
自引率
8.20%
发文量
1375
审稿时长
2.3 months
期刊介绍:
The Journal of Agricultural and Food Chemistry publishes high-quality, cutting edge original research representing complete studies and research advances dealing with the chemistry and biochemistry of agriculture and food. The Journal also encourages papers with chemistry and/or biochemistry as a major component combined with biological/sensory/nutritional/toxicological evaluation related to agriculture and/or food.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


