核磁共振分析:一个半自动化核磁共振代谢物分析的开源Web应用程序
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
虽然核磁共振(NMR)波谱的数据采集和初始信号预处理已经实现了高度自动化,但下游处理──特别是谱的剖面──仍是整个核磁共振分析工作流程的瓶颈。已经做出了一些努力来缓解这一瓶颈,但这些解决方案往往以自动化的增加为代价,在其他方面受到限制。在这项工作中,我们介绍了nmRanalysis,这是一个用户友好的web应用程序,它集成了现有分析工具的优势,以实现更自动化的分析工作流。核磁共振分析还结合了新的功能,包括用于代谢物鉴定的机器学习驱动的推荐系统,进一步提高了核磁共振分析相对于它所结合的单个工具的实用性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
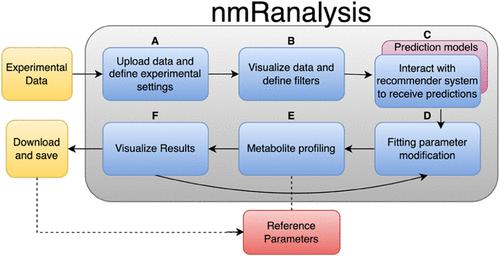
nmRanalysis: An Open-Source Web Application for Semi-automated NMR Metabolite Profiling
Though data acquisition and initial signal preprocessing of nuclear magnetic resonance (NMR) spectra have achieved high degrees of automation, downstream processing─specifically the profiling of spectra─has bottlenecked the overall NMR analysis workflow. Several efforts have been made to mitigate this bottleneck, but these solutions often trade an increase in automation for limitations elsewhere. In this work, we introduce nmRanalysis, a user-friendly web application that integrates the strengths of existing profiling tools for a more automated profiling workflow. nmRanalysis additionally incorporates novel features, including a machine-learning-driven recommender system for metabolite identification, further increasing the utility of nmRanalysis over the individual tools that it incorporates.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


