发现乳腺癌多倍体巨细胞新抑制剂的高通量经验和虚拟筛选
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
乳腺癌的治疗耐药越来越多地归因于多倍体巨型癌细胞(pgcc),它们通过全基因组加倍产生,对标准治疗表现出更高的适应性。这些细胞的特点是细胞核增大,DNA含量增加,在治疗压力下往往处于休眠状态,导致疾病复发。尽管它们在耐药性中起着关键作用,但有效靶向pgcc的策略有限,这主要是由于缺乏高通量方法来评估它们的生存能力。传统的检测方法缺乏检测pgcc特异性消除所需的灵敏度,这促使了新方法的发展。为了应对这一挑战,我们开发了一种高通量单细胞形态分析工作流程,旨在区分选择性抑制非pgcc、pgcc或两者的化合物。使用这种方法,我们筛选了2726种FDA批准的1期药物库,确定了有希望的抗pgcc候选药物,包括蛋白酶体抑制剂、FOXM1、CHK和大环内酯。值得注意的是,用大环内酯吡啶处理的细胞的RNA-Seq分析显示,AXL抑制是靶向pgcc的潜在策略。虽然我们的单细胞形态分析管道是强大的,所有现有的化合物的经验测试是不切实际和低效的。为了克服这一限制,我们训练了一个机器学习模型来预测抗pgcc的有效性,整合了化学指纹和来自先前出版物和数据库的化合物描述。该模型与实验结果高度相关,并预测了超过6000种药物库中的有效化合物。在排名靠前的预测中,我们通过细胞系和患者衍生模型实验验证了五种化合物是有效的PGCC抑制剂。这些发现强调了将高通量经验筛选与基于机器学习的虚拟筛选相结合的协同潜力,以加速新疗法的发现,特别是针对乳腺癌治疗耐药的pgcc。本文章由计算机程序翻译,如有差异,请以英文原文为准。
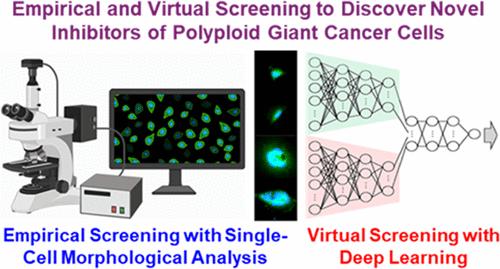
High-Throughput Empirical and Virtual Screening To Discover Novel Inhibitors of Polyploid Giant Cancer Cells in Breast Cancer
Therapy resistance in breast cancer is increasingly attributed to polyploid giant cancer cells (PGCCs), which arise through whole genome doubling and exhibit heightened resilience to standard treatments. Characterized by enlarged nuclei and increased DNA content, these cells tend to be dormant under therapeutic stress, driving disease relapse. Despite their critical role in resistance, strategies to effectively target PGCCs are limited, largely due to the lack of high-throughput methods for assessing their viability. Traditional assays lack the sensitivity needed to detect PGCC-specific elimination, prompting the development of novel approaches. To address this challenge, we developed a high-throughput single-cell morphological analysis workflow designed to differentiate compounds that selectively inhibit non-PGCCs, PGCCs, or both. Using this method, we screened a library of 2726 FDA Phase 1-approved drugs, identifying promising anti-PGCC candidates, including proteasome inhibitors, FOXM1, CHK, and macrocyclic lactones. Notably, RNA-Seq analysis of cells treated with the macrocyclic lactone Pyronaridine revealed AXL inhibition as a potential strategy for targeting PGCCs. Although our single-cell morphological analysis pipeline is powerful, empirical testing of all existing compounds is impractical and inefficient. To overcome this limitation, we trained a machine learning model to predict anti-PGCC efficacy in silico, integrating chemical fingerprints and compound descriptions from prior publications and databases. The model demonstrated a high correlation with experimental outcomes and predicted efficacious compounds in an expanded library of over 6,000 drugs. Among the top-ranked predictions, we experimentally validated five compounds as potent PGCC inhibitors using cell lines and patient-derived models. These findings underscore the synergistic potential of integrating high-throughput empirical screening with machine learning-based virtual screening to accelerate the discovery of novel therapies, particularly for targeting therapy-resistant PGCCs in breast cancer.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


