通过重复元素之间的工程重组来随机化人类基因组
IF 45.8
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
我们缺乏必要的工具来编辑DNA序列,以研究99%的非编码人类基因组。为了解决这一差距,我们应用CRISPR引物编辑将重组手柄插入到重复序列中,每个细胞系多达1697个,从而能够产生大规模的缺失、倒位、易位和环状DNA。重组酶诱导在每个细胞中产生100多个随机的兆酶大小的重排。随着时间的推移,我们追踪了这些重排,以测量选择压力,发现人们更倾向于避免必要基因的较短变异。我们对29个具有多重重排的克隆进行了表征,发现缺失对变体中基因的表达有影响,但对附近基因没有影响。这种基因组混乱策略使得大量缺失、序列重定位和调控元件的插入能够探索基因组的可有可无和组织。本文章由计算机程序翻译,如有差异,请以英文原文为准。
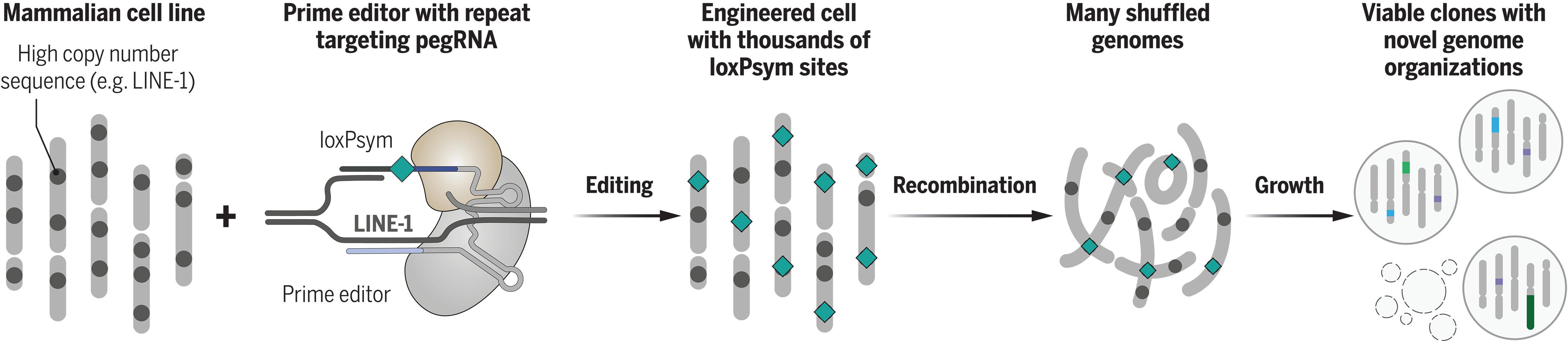
Randomizing the human genome by engineering recombination between repeat elements
We lack tools to edit DNA sequences at scales necessary to study 99% of the human genome that is noncoding. To address this gap, we applied CRISPR prime editing to insert recombination handles into repetitive sequences, up to 1697 per cell line, which enables generating large-scale deletions, inversions, translocations, and circular DNA. Recombinase induction produced more than 100 stochastic megabase-sized rearrangements in each cell. We tracked these rearrangements over time to measure selection pressures, finding a preference for shorter variants that avoided essential genes. We characterized 29 clones with multiple rearrangements, finding an impact of deletions on expression of genes in the variant but not on nearby genes. This genome-scrambling strategy enables large deletions, sequence relocations, and the insertion of regulatory elements to explore genome dispensability and organization.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science
综合性期刊-综合性期刊
CiteScore
61.10
自引率
0.90%
发文量
0
审稿时长
2.1 months
期刊介绍:
Science is a leading outlet for scientific news, commentary, and cutting-edge research. Through its print and online incarnations, Science reaches an estimated worldwide readership of more than one million. Science’s authorship is global too, and its articles consistently rank among the world's most cited research.
Science serves as a forum for discussion of important issues related to the advancement of science by publishing material on which a consensus has been reached as well as including the presentation of minority or conflicting points of view. Accordingly, all articles published in Science—including editorials, news and comment, and book reviews—are signed and reflect the individual views of the authors and not official points of view adopted by AAAS or the institutions with which the authors are affiliated.
Science seeks to publish those papers that are most influential in their fields or across fields and that will significantly advance scientific understanding. Selected papers should present novel and broadly important data, syntheses, or concepts. They should merit recognition by the wider scientific community and general public provided by publication in Science, beyond that provided by specialty journals. Science welcomes submissions from all fields of science and from any source. The editors are committed to the prompt evaluation and publication of submitted papers while upholding high standards that support reproducibility of published research. Science is published weekly; selected papers are published online ahead of print.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


