磁珠上深度学习辅助数字荧光免疫分析法超灵敏测定蛋白质生物标志物
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
基于随机分散磁珠(MBs)的数字荧光免疫分析法(DFI)是超灵敏测定蛋白质生物标志物的有力方法之一。然而,在DFI中,由于信号与背景的比率和手动计数珠子的速度较低,因此提高检测极限(LOD)是具有挑战性的。在此,我们利用UNet3+框架内的一种新的混合注意力机制开发了一个深度学习网络(ATTBeadNet),用于准确快速地计数mb,并提出了一个使用窄峰和光学稳定性的CdS量子点(QDs)的DFI,这是首次报道。应用开发的ATTBeadNet对MBs进行计数,F1评分(95.91%)高于其他方法(ImageJ, 68.33%;基于计算机视觉的,92.99%;全卷积网络,75.00%;基于掩模区域的卷积神经网络(70.34%)。在证明原理的基础上,提出了一种基于夹心mb的DFI,其中以人白细胞介素-6 (IL-6)作为模型蛋白生物标志物,以抗体结合的链亲和素包被的mb作为捕获mb,以抗体- hrp -酪酰胺功能化的CdS量子点作为结合报告基因。将所开发的ATTBeadNet应用于基于mb的IL-6 (20 μL)的DFI,线性范围为5 ~ 100 fM, LOD为3.1 fM,优于ImageJ方法(线性范围为30 ~ 100 fM, LOD为20 fM)。这项工作表明,深度学习网络与DFI的集成是一种高灵敏度和准确测定蛋白质生物标志物的有前途的策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。
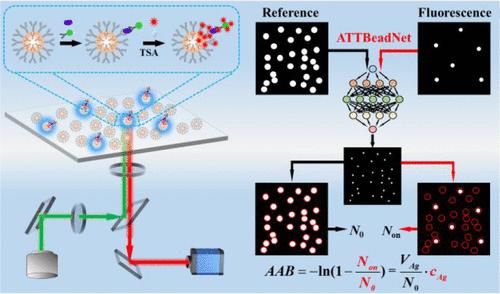
Deep-Learning-Assisted Digital Fluorescence Immunoassay on Magnetic Beads for Ultrasensitive Determination of Protein Biomarkers
Digital fluorescence immunoassay (DFI) based on random dispersion magnetic beads (MBs) is one of the powerful methods for ultrasensitive determination of protein biomarkers. However, in the DFI, improving the limit of detection (LOD) is challenging since the ratio of signal-to-background and the speed of manual counting beads are low. Herein, we developed a deep-learning network (ATTBeadNet) by utilizing a new hybrid attention mechanism within a UNet3+ framework for accurately and fast counting the MBs and proposed a DFI using CdS quantum dots (QDs) with narrow peak and optical stability as reported at first time. The developed ATTBeadNet was applied to counting the MBs, resulting in the F1 score (95.91%) being higher than those of other methods (ImageJ, 68.33%; computer vision-based, 92.99%; fully convolutional network, 75.00%; mask region-based convolutional neural network, 70.34%). On principle-on-proof, a sandwich MB-based DFI was proposed, in which human interleukin-6 (IL-6) was taken as a model protein biomarker, while antibody-bound streptavidin-coated MBs were used as capture MBs and antibody-HRP-tyramide-functionalized CdS QDs were used as the binding reporter. When the developed ATTBeadNet was applied to the MB-based DFI of IL-6 (20 μL), the linear range from 5 to 100 fM and an LOD of 3.1 fM were achieved, which are better than those using the ImageJ method (linear range from 30 to 100 fM and LOD of 20 fM). This work demonstrates that the integration of the deep-learning network with DFI is a promising strategy for the highly sensitive and accurate determination of protein biomarkers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


