使用可富集连接子的定量聚糖-蛋白质交联质谱揭示了广泛的聚糖介导的蛋白质相互作用网络
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
细胞膜中的蛋白质-蛋白质相互作用通常由聚糖介导,末端唾液酸通常参与这些相互作用。为了探究相互作用的本质,我们开发了涉及糖蛋白聚糖和蛋白质多肽部分的定量交联方法。我们设计并合成了生物素化的可富集交联剂,这些交联剂被点击标记,以代谢方式将叠氮-唾液酸结合到细胞表面聚糖上,从而允许叠氮-聚糖与近端多肽上的赖氨酸残基交联。通过链霉亲和素树脂珠亲和纯化,用生物素基团富集糖肽-肽交联(GPx)。工作流程使用两个连接,一个光可切割和另一个二硫,开发并应用于揭示唾液酸介导的PNT2(前列腺)细胞表面蛋白网络。鉴定了糖肽-肽对,其中12000个GPx通过唾液化糖形式连接,揭示了源糖蛋白与近700个靶蛋白之间的相互作用。蛋白质之间的相互作用由多达40对肽对表征,蛋白质之间的相互作用程度由GPx的数量来优先排序。通过计数鉴定蛋白对的GPx数量进行定量。大量的膜蛋白,如ITGB1,产生了一个由大约400个其他蛋白质组成的相互作用组,这些蛋白质从相互作用最广泛,GPx数量最多,到至少一个。相互作用组通过已发表的数据库进一步单独确认。该工具将增强我们对蛋白质-蛋白质相互作用的糖基化的理解,并为治疗提供新的潜在靶点。本文章由计算机程序翻译,如有差异,请以英文原文为准。
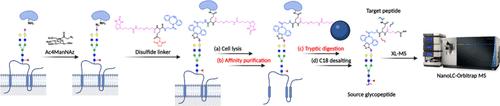
Quantitative Glycan-Protein Cross-Linking Mass Spectrometry Using Enrichable Linkers Reveals Extensive Glycan-Mediated Protein Interaction Networks
Protein–protein interactions in the cell membrane are typically mediated by glycans, with terminal sialic acid often involved in these interactions. To probe the nature of the interactions, we developed quantitative cross-linking methods involving the glycans of the glycoproteins and the polypeptide moieties of proteins. We designed and synthesized biotinylated enrichable cross-linkers that were click-tagged to metabolically incorporate azido-sialic acid on cell surface glycans to allow cross-linking of the azido-glycans with lysine residues on proximal polypeptides. The glycopeptide–peptide cross-links (GPx) were enriched using biotin groups through affinity purification with streptavidin resin beads. Workflows using two linkers, one photocleavable and the other disulfide, were developed and applied to reveal the sialic acid-mediated cell-surface protein networks of PNT2 (prostate) cells. Glycopeptide–peptide pairs were identified, with 12000 GPx linked by sialylated glycoforms revealing interactions between source glycoproteins and nearly 700 target proteins. Protein–protein interactions were characterized by as many as 40 peptide pairs, and the extent of the interactions between proteins was prioritized by the number of GPx. Quantitation was performed by counting the number of GPx that identify the protein pairs. Abundant membrane proteins such as ITGB1 yielded an interactome consisting of around 400 other proteins, which were ranked from the most extensive interaction, having the largest number of GPx, to at least one. The interactome was further confirmed separately by published databases. This tool will enhance our understanding of glycosylation on protein–protein interactions and provide new potential targets for therapeutics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


