多段注射-毛细管电泳-质谱法快速自动预处理大规模血清代谢组学数据的软件工具
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
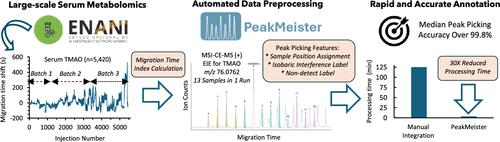
在分析复杂的生物标本时,基于质谱(MS)的代谢组学通常依赖于分离技术,以提高方法分辨率、代谢组学覆盖范围、定量性能和/或未知鉴定。然而,低样本吞吐量和复杂的数据预处理程序仍然是可负担得起的代谢组学研究的主要障碍,这些研究可扩展到大量人群。在此,我们在R统计环境中引入了PeakMeister作为一种新的软件工具,可以对通过多段注射-毛细管电泳-质谱(MSI-CE-MS)获得的血清代谢组学数据进行标准化处理,这是一个高通量分离平台(<;4分钟/样品),利用了在单个分析运行中连续注射13个样品的格式。我们对PeakMeister进行了严格的验证,分析了在巴西国家儿童营养调查(ENANI-2019)的5000份血清和420份质量控制样本中一致测量的47种离子代谢物,包括在8个月的时间内,在40天内分三批共获得的224,983个代谢物峰。引入了一个使用11个内部标准的迁移时间指数来纠正迁移时间的大变化,这允许可靠的峰注释、峰整合和具有两个侧翼内部标准或单个稳定同位素内部标准的血清代谢物的样品位置分配。与经验丰富的分析师使用供应商软件手动处理MSI-CE-MS数据相比,PeakMeister将数据预处理时间缩短了30倍,同时还实现了出色的峰注释保真度(中位数精度>;99.9%),可接受的中间精度(中位数CV = 16.0%),一致的代谢物峰整合(平均偏差= - 2.1%),并且在量化NIST SRM-1950的16种血浆代谢物时具有良好的一致性(平均偏差= - 1.3%)。ENANI-2019研究还报告了巴西5岁以下儿童全国营养调查中40种血清代谢物的参考范围。MSI-CE-MS与PeakMeister相结合,可以快速和自动化地处理大规模代谢组学研究,这些研究可以容忍非线性迁移时移,而无需复杂的动态时间扭曲或有效的迁移尺度转换。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A Software Tool for Rapid and Automated Preprocessing of Large-Scale Serum Metabolomic Data by Multisegment Injection-Capillary Electrophoresis-Mass Spectrometry
Mass spectrometry (MS)-based metabolomics often rely on separation techniques when analyzing complex biological specimens to improve method resolution, metabolome coverage, quantitative performance, and/or unknown identification. However, low sample throughput and complicated data preprocessing procedures remain major barriers to affordable metabolomic studies that are scalable to large populations. Herein, we introduce PeakMeister as a new software tool in the R statistical environment to enable standardized processing of serum metabolomic data acquired by multisegment injection-capillary electrophoresis-mass spectrometry (MSI-CE-MS), a high-throughput separation platform (<4 min/sample) which takes advantage of a serial injection format of 13 samples within a single analytical run. We performed a rigorous validation of PeakMeister by analyzing 47 cationic metabolites consistently measured in 5,000 serum and 420 quality control samples from the Brazilian National Survey on Child Nutrition (ENANI-2019) comprising a total of 224,983 metabolite peaks acquired in 40 days across three batches over an eight-month period. A migration time index using a panel of 11 internal standards was introduced to correct for large variations in migration times, which allowed for reliable peak annotation, peak integration, and sample position assignment for serum metabolites having two flanking internal standards or a single comigrating stable-isotope internal standard. PeakMeister accelerated data preprocessing times by 30-fold compared to manual processing of MSI-CE-MS data by an experienced analyst using vendor software, while also achieving excellent peak annotation fidelity (median accuracy >99.9%), acceptable intermediate precision (median CV = 16.0%), consistent metabolite peak integration (mean bias = −2.1%), and good mutual agreement when quantifying 16 plasma metabolites from NIST SRM-1950 (mean bias = −1.3%). Reference ranges are also reported for 40 serum metabolites in a national nutritional survey of Brazilian children under 5 years of age from the ENANI-2019 study. MSI-CE-MS in conjunction with PeakMeister allows for rapid and automated processing of large-scale metabolomic studies that tolerate nonlinear migration time shifts without complicated dynamic time warping or effective mobility scale transformations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


