蛋白质组学标准计划 光谱库和碎片离子峰注释的标准格式:mzSpecLib 和 mzPAF
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
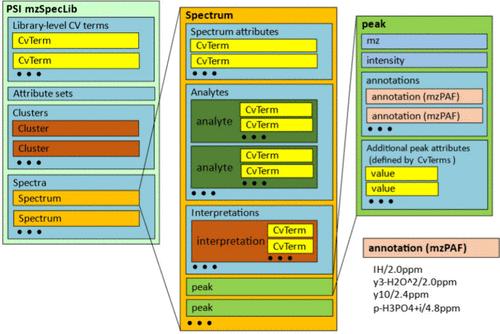
质谱库是参考光谱的集合,通常与生成光谱的特定分析物相关,用于对新光谱进行进一步的下游分析。用于编码质谱库的格式多种多样,但没有一种格式经过标准化处理,以确保广泛适用于多种应用。作为人类蛋白质组组织蛋白质组学标准倡议(PSI)的一部分,我们开发了一种用于编码谱库的标准化格式,称为 mzSpecLib (https://psidev.info/mzSpecLib)。它主要是一种数据模型,可使用可扩展的 PSI-MS 受控词汇表灵活编码库条目元数据,并可在不同的序列化格式中编码和转换。我们还为片段离子峰注释开发了一种标准化数据模型和序列化格式,称为 mzPAF (https://psidev.info/mzPAF)。它被定义为一个单独的标准,因为除了光谱库之外,它还可用于其他应用。mzSpecLib 和 mzPAF 标准与现有的 PSI 标准兼容,如 ProForma 2.0 和通用光谱标识符。mzSpecLib 和 mzPAF 标准主要是为蛋白质组学应用中的肽定义的,并提供基本的小分子支持。今后,它们还可扩展到需要为非肽类分析物编码谱库的其他领域。本文章由计算机程序翻译,如有差异,请以英文原文为准。
The Proteomics Standards Initiative Standardized Formats for Spectral Libraries and Fragment Ion Peak Annotations: mzSpecLib and mzPAF
Mass spectral libraries are collections of reference spectra, usually associated with specific analytes from which the spectra were generated, that are used for further downstream analysis of new spectra. There are many different formats used for encoding spectral libraries, but none have undergone a standardization process to ensure broad applicability to many applications. As part of the Human Proteome Organization Proteomics Standards Initiative (PSI), we have developed a standardized format for encoding spectral libraries, called mzSpecLib (https://psidev.info/mzSpecLib). It is primarily a data model that flexibly encodes metadata about the library entries using the extensible PSI-MS controlled vocabulary and can be encoded in and converted between different serialization formats. We have also developed a standardized data model and serialization for fragment ion peak annotations, called mzPAF (https://psidev.info/mzPAF). It is defined as a separate standard, since it may be used for other applications besides spectral libraries. The mzSpecLib and mzPAF standards are compatible with existing PSI standards such as ProForma 2.0 and the Universal Spectrum Identifier. The mzSpecLib and mzPAF standards have been primarily defined for peptides in proteomics applications with basic small molecule support. They could be extended in the future to other fields that need to encode spectral libraries for nonpeptidic analytes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


