NMRMind: A Transformer-Based Model Enabling the Elucidation from Multidimensional NMR to Structures
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Nuclear magnetic resonance (NMR) data provides rich quantum information on molecular structure, which is closely related to chemical structure and widely used for structural characterization in chemical discovery. Despite substantial advances in spectral analysis techniques, few existing models have demonstrated satisfactory performance in accurate NMR interpretation. Herein, we introduce NMRMind, a Transformer-based generative framework that directly elucidates molecular structures from NMR spectral data. NMRMind was pretrained on a data set comprising 45 million 1D NMR spectra and subsequently fine-tuned on a self-curated benchmark consisting of 2.2 million 1D and 2D NMR spectra. Using a mixed-modality dropout strategy during training, NMRMind achieved excellent performance, attaining a Top-1 accuracy of 92.07% across all input conditions on the structure elucidation task with a speed of <0.05 s per elucidation. Additionally, NMRMind maintained a Top-1 accuracy of 85.10% when only one-dimensional and two-dimensional NMR data were used as input, without considering molecular formulas or fragments. Moreover, the application of NMRMind facilitated the discovery of six previously uncharacterized natural products from Magnolia officinalis and successfully elucidated the structures of six unexpected products resulting from synthetic reactions, thereby expanding the accessible chemical space and providing novel insights into chemical mechanisms. These results demonstrate that NMRMind is a powerful and generalizable platform for chemistry research.
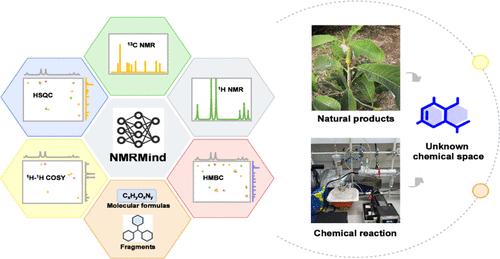
核磁共振思维:一个基于变压器的模型,实现了从多维核磁共振到结构的解析
核磁共振(NMR)数据提供了丰富的分子结构量子信息,与化学结构密切相关,广泛用于化学发现中的结构表征。尽管光谱分析技术取得了长足的进步,但很少有现有的模型在精确的核磁共振解释中表现出令人满意的性能。在这里,我们介绍了NMRMind,一个基于变压器的生成框架,可以直接从核磁共振光谱数据中阐明分子结构。NMRMind在包含4500万张1D NMR波谱的数据集上进行了预训练,随后在包含220万张1D和2D NMR波谱的自策划基准上进行了微调。在训练过程中使用混合模态退出策略,NMRMind在所有输入条件下都达到了92.07%的Top-1准确率,每个解析速度为0.05 s。此外,当仅使用一维和二维核磁共振数据作为输入,不考虑分子式或片段时,NMRMind保持了85.10%的Top-1准确率。此外,NMRMind的应用促进了厚朴六种以前未被表征的天然产物的发现,并成功阐明了合成反应产生的六种意想不到的产物的结构,从而扩大了可访问的化学空间,并为化学机制提供了新的见解。这些结果表明,NMRMind是一个强大的、可推广的化学研究平台。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


