A phylogenetic perspective on the performance of Lactococcus lactis as starter culture in milk and plant milks
IF 8
1区 农林科学
Q1 FOOD SCIENCE & TECHNOLOGY
引用次数: 0
Abstract
Lactococcus lactis is used as a starter culture in dairy fermentations but also occurs in spontaneous plant fermentations. Phylogenetic analyses have differentiated Lc. lactis into distinct clades. One clade consists exclusively of dairy isolates, which evolved from a more ancestral clade comprising both plant and dairy isolates. However, it remains unclear whether strains from these clades differ in their ability to ferment milk and plant-based milk. This study aimed to compare the fermentative properties of Lc. lactis from a phylogenetic perspective. A core genome phylogenetic tree based on high-quality Lc. lactis genomes revealed distinct ancestral, plant, and dairy lineages which were not congruent with the current subspecies classification. The phylogeny of the dairy lineage and the geographic origin of the isolates supported domestication of the dairy lineage. A phenotypic analysis of 12 representative strains from the lineages (ancestral, plant and dairy) demonstrated that sugar metabolism and acidification profiles aligned with genotypic differences. Dairy-lineage strains acidified bovine milk more effectively and produced higher levels of lactic acid. In contrast, most strains of the plant-lineage were unable to acidify bovine milk below pH 5.4, or to produce lactic acid in milk. These strains, however, exhibited efficient metabolism of raffinose family oligosaccharides and rapidly acidified lupin milk. Lc. lactis FUA3579, classified within the ancestral lineage, effectively acidified both plant-based and dairy matrices and metabolized sugars in both. This study highlights the metabolic divergence among Lc. lactis lineages and provides insights for selection of starter cultures tailored for milk or plant-based dairy alternatives.
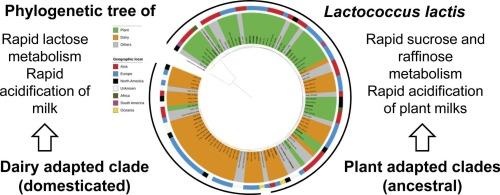
乳球菌在牛奶和植物乳中作为发酵剂性能的系统发育研究
乳酸乳球菌在乳制品发酵中用作发酵剂,但也发生在植物自发发酵中。系统发育分析已分化出Lc。成明显分支的冰川。一个分支完全由乳制品分离株组成,它从一个更古老的分支进化而来,包括植物和乳制品分离株。然而,目前尚不清楚这些分支的菌株在发酵牛奶和植物奶的能力上是否存在差异。本研究旨在比较Lc的发酵特性。从系统发育的角度看乳石。基于高质量Lc的核心基因组系统发育树。乳酸菌基因组显示了不同的祖先、植物和乳系谱系,这与目前的亚种分类不一致。奶牛谱系的系统发育和分离株的地理起源支持了奶牛谱系的驯化。对来自祖先、植物和奶牛谱系的12个代表性菌株的表型分析表明,糖代谢和酸化谱与基因型差异一致。奶牛系菌株更有效地酸化了牛奶,并产生了更高水平的乳酸。相比之下,大多数植物系菌株不能使pH值低于5.4的牛奶酸化,也不能在牛奶中产生乳酸。然而,这些菌株对棉子糖家族低聚糖具有高效的代谢和快速酸化的罗苹乳。信用证。laactis FUA3579属于祖先谱系,可有效酸化植物基和乳制品基质,并代谢两者中的糖。本研究突出了Lc之间的代谢差异。乳酸菌谱系,并为选择适合牛奶或植物性乳制品替代品的发酵剂提供见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Food Research International
工程技术-食品科技
CiteScore
12.50
自引率
7.40%
发文量
1183
审稿时长
79 days
期刊介绍:
Food Research International serves as a rapid dissemination platform for significant and impactful research in food science, technology, engineering, and nutrition. The journal focuses on publishing novel, high-quality, and high-impact review papers, original research papers, and letters to the editors across various disciplines in the science and technology of food. Additionally, it follows a policy of publishing special issues on topical and emergent subjects in food research or related areas. Selected, peer-reviewed papers from scientific meetings, workshops, and conferences on the science, technology, and engineering of foods are also featured in special issues.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


