Systems Age: a single blood methylation test to quantify aging heterogeneity across 11 physiological systems
IF 19.4
Q1 CELL BIOLOGY
引用次数: 0
Abstract
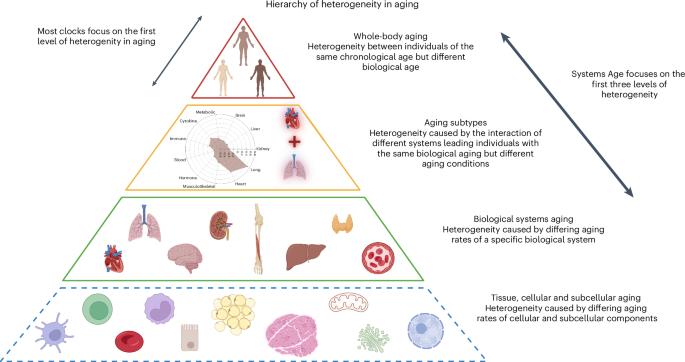
Aging occurs at different rates across individuals and physiological systems, but most epigenetic clocks provide a single age estimate, overlooking within-person variation. Here we developed systems-based DNA methylation clocks that measure aging in 11 distinct physiological systems—Heart, Lung, Kidney, Liver, Brain, Immune, Inflammatory, Blood, Musculoskeletal, Hormone and Metabolic—using data from a single blood draw. By integrating supervised and unsupervised machine learning with clinical biomarkers, functional assessments and mortality risk, we derived system-specific scores that outperformed existing global clocks in predicting relevant diseases and aging phenotypes. We also created a composite Systems Age score to capture overall multisystem aging. Clustering individuals based on these scores revealed distinct biological aging subtypes, each associated with unique patterns of health decline and disease risk. This framework enables a more granular and clinically relevant assessment of biological aging and may support personalized approaches to monitor and target system-specific aging processes. Sehgal et al. report Systems Age as a framework to capture within-person heterogeneity in aging using a single blood-based epigenetic assay that measures aging across 11 body systems and identifies aging subtypes, enabling personalized prediction of disease risk and tailoring of longevity interventions.
系统年龄:单一血液甲基化测试来量化11个生理系统的衰老异质性。
不同个体和生理系统的衰老速度不同,但大多数表观遗传时钟提供单一的年龄估计,忽略了人体内的变化。在这里,我们开发了基于系统的DNA甲基化时钟,可以测量11个不同生理系统的衰老——心脏、肺、肾、肝、脑、免疫、炎症、血液、肌肉骨骼、激素和代谢——使用单次抽血的数据。通过将有监督和无监督机器学习与临床生物标志物、功能评估和死亡风险相结合,我们得出了在预测相关疾病和衰老表型方面优于现有全球时钟的系统特异性评分。我们还创建了一个复合系统年龄评分来捕获整个多系统老化。基于这些分数的个体聚类揭示了不同的生物衰老亚型,每种亚型都与健康衰退和疾病风险的独特模式相关。该框架能够对生物衰老进行更细致和临床相关的评估,并可能支持个性化方法来监测和靶向系统特异性衰老过程。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


