Machine learning–based penetrance of genetic variants
IF 45.8
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Accurate variant penetrance estimation is crucial for precision medicine. We constructed machine learning (ML) models for 10 diseases using 1,347,298 participants with electronic health records, then applied them to an independent cohort with linked exome data. Resulting probabilities were used to evaluate ML penetrance of 1648 rare variants in 31 autosomal dominant disease-predisposition genes. ML penetrance was variable across variant classes, but highest for pathogenic and loss-of-function variants, and was associated with clinical outcomes and functional data. Compared with conventional case-versus-control approaches, ML penetrance provided refined quantitative estimates and aided the interpretation of variants of uncertain significance and loss-of-function variants by delineating clinical trajectories over time. By leveraging ML and deep phenotyping, we present a scalable approach to accurately quantify disease risk of variants.
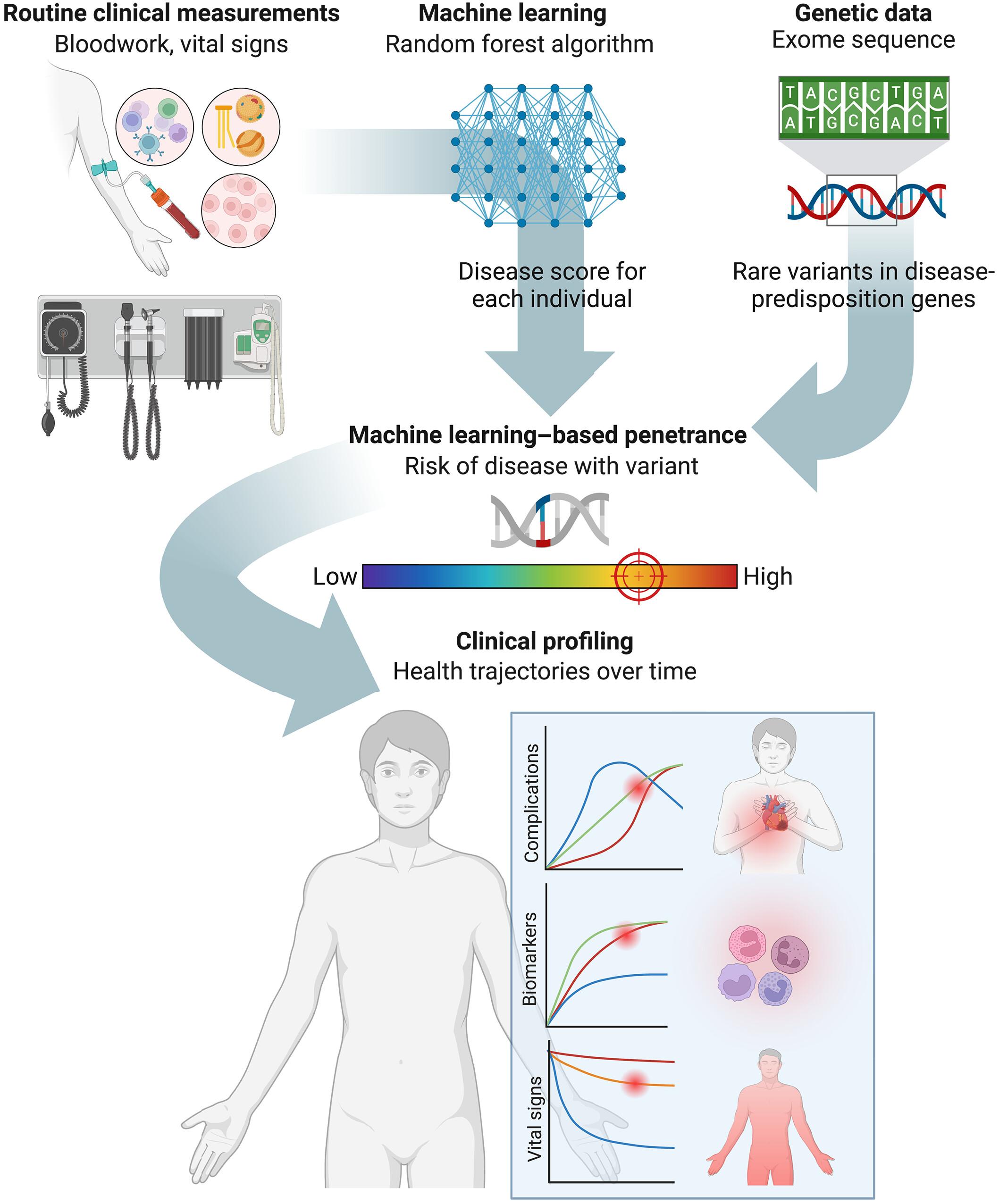
基于机器学习的基因变异外显率
准确的变异外显率估计对精准医疗至关重要。我们使用1,347,298名具有电子健康记录的参与者构建了10种疾病的机器学习(ML)模型,然后将其应用于具有关联外显子组数据的独立队列。结果概率用于评估31个常染色体显性疾病易感基因中1648个罕见变异的ML外显率。ML外显率在不同的变异类别中是可变的,但在致病性和功能丧失变异中最高,并且与临床结果和功能数据相关。与传统的病例对照方法相比,ML外显率提供了精确的定量估计,并通过描述临床轨迹来帮助解释不确定意义的变异和功能丧失的变异。通过利用机器学习和深度表型,我们提出了一种可扩展的方法来准确量化变异的疾病风险。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Science
综合性期刊-综合性期刊
CiteScore
61.10
自引率
0.90%
发文量
0
审稿时长
2.1 months
期刊介绍:
Science is a leading outlet for scientific news, commentary, and cutting-edge research. Through its print and online incarnations, Science reaches an estimated worldwide readership of more than one million. Science’s authorship is global too, and its articles consistently rank among the world's most cited research.
Science serves as a forum for discussion of important issues related to the advancement of science by publishing material on which a consensus has been reached as well as including the presentation of minority or conflicting points of view. Accordingly, all articles published in Science—including editorials, news and comment, and book reviews—are signed and reflect the individual views of the authors and not official points of view adopted by AAAS or the institutions with which the authors are affiliated.
Science seeks to publish those papers that are most influential in their fields or across fields and that will significantly advance scientific understanding. Selected papers should present novel and broadly important data, syntheses, or concepts. They should merit recognition by the wider scientific community and general public provided by publication in Science, beyond that provided by specialty journals. Science welcomes submissions from all fields of science and from any source. The editors are committed to the prompt evaluation and publication of submitted papers while upholding high standards that support reproducibility of published research. Science is published weekly; selected papers are published online ahead of print.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


