SST-DUNet: Smart Swin Transformer and Dense UNet for automated preclinical fMRI skull stripping
IF 2.3
4区 医学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Background:
Skull stripping is a common preprocessing step in Magnetic Resonance Imaging (MRI) pipelines and is often performed manually. Automating this process is challenging for preclinical data due to variations in brain geometry, resolution, and tissue contrast. Existing methods for MRI skull stripping often struggle with the low resolution and varying slice sizes found in preclinical functional MRI (fMRI) data.
New method:
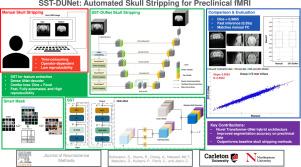
This study proposes a novel method that integrates a Dense UNet-based architecture with a feature extractor based on the Smart Swin Transformer (SST), called SST-DUNet. The Smart Shifted Window Multi-Head Self-Attention (SSW-MSA) module in SST replaces the mask-based module in the Swin Transformer (ST), enabling the learning of distinct channel-wise features while focusing on relevant dependencies within brain structures. This modification allows the model to better handle the complexities of fMRI skull stripping, such as low resolution and variable slice sizes. To address class imbalance in preclinical data, a combined loss function using Focal and Dice loss is applied.
Results:
The model was trained on rat fMRI images and evaluated across three in-house datasets, achieving Dice similarity scores of 98.65%, 97.86%, and 98.04%.
Comparison with existing methods:
We compared our method with conventional and deep learning-based approaches, demonstrating its superiority over state-of-the-art methods.
Conclusions:
The fMRI results using SST-DUNet closely align with those from manual skull stripping for both seed-based and independent component analyses, indicating that SST-DUNet can effectively substitute manual brain extraction in rat fMRI analysis.
SST-DUNet:用于自动临床前fMRI颅骨剥离的智能Swin变压器和密集UNet
背景:颅骨剥离是磁共振成像(MRI)管道中常见的预处理步骤,通常手工进行。由于大脑几何形状、分辨率和组织对比度的变化,自动化这一过程对临床前数据具有挑战性。现有的MRI颅骨剥离方法经常与临床前功能MRI (fMRI)数据中发现的低分辨率和不同的切片大小作斗争。新方法:本研究提出了一种新方法,将基于Dense unet的架构与基于Smart Swin Transformer (SST)的特征提取器(称为SST- dunet)集成在一起。SST中的Smart shift Window Multi-Head Self-Attention (SSW-MSA)模块取代了Swin Transformer (ST)中基于掩模的模块,能够学习不同的通道智能特征,同时专注于大脑结构中的相关依赖关系。这种修改使模型能够更好地处理fMRI颅骨剥离的复杂性,例如低分辨率和可变切片大小。为了解决临床前数据中的类别不平衡,应用了一个使用Focal和Dice损失的组合损失函数。结果:该模型在大鼠fMRI图像上进行了训练,并在三个内部数据集上进行了评估,获得了98.65%、97.86%和98.04%的Dice相似性得分。与现有方法的比较:我们将我们的方法与传统和基于深度学习的方法进行了比较,证明了其优于最先进的方法。结论:SST-DUNet的fMRI结果与手工颅骨剥离的结果在种子分析和独立成分分析中都非常接近,表明SST-DUNet可以有效地替代手工脑提取进行大鼠fMRI分析。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Neuroscience Methods
医学-神经科学
CiteScore
7.10
自引率
3.30%
发文量
226
审稿时长
52 days
期刊介绍:
The Journal of Neuroscience Methods publishes papers that describe new methods that are specifically for neuroscience research conducted in invertebrates, vertebrates or in man. Major methodological improvements or important refinements of established neuroscience methods are also considered for publication. The Journal''s Scope includes all aspects of contemporary neuroscience research, including anatomical, behavioural, biochemical, cellular, computational, molecular, invasive and non-invasive imaging, optogenetic, and physiological research investigations.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


