Refining the generation, interpretation and application of multi-organ, multi-omics biological aging clocks
IF 19.4
Q1 CELL BIOLOGY
引用次数: 0
Abstract
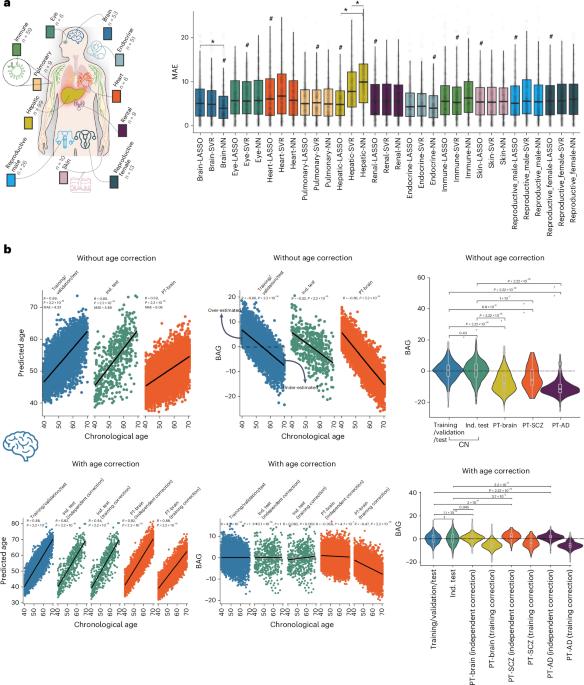
Multi-organ biological aging clocks derived from clinical phenotypes and neuroimaging data have emerged as valuable tools for studying human aging and disease. Plasma proteomics provides an additional molecular dimension to enrich these clocks. In this study, I developed 11 multi-organ proteome-based biological age gaps (ProtBAGs) using 2,448 plasma proteins from 43,498 participants in the UK Biobank. Here I highlight methodological and clinical considerations for developing and using these clocks, including correction for age bias, organ specificity of proteins, sample size and underlying pathologies in the training data, which can affect model generalizability and clinical interpretability. In addition, I integrated 11 ProtBAGs with previously developed nine multi-organ phenotype-based biological age gaps to investigate genetic overlap and causal associations with disease endpoints. Finally, I show that incorporating features across organs improves predictions for systemic disease categories and all-cause mortality. These analyses provide methodological and clinical insights for developing and interpreting these clocks and highlight future avenues toward a multi-organ, multi-omics biological aging clock framework. In developing multi-organ-based and multi-omics-based biological aging clocks and linking them to underlying genetics and clinical phenotypes, Wen demonstrates the value of methodological and clinical considerations for enhancing model generalizability and clinical interpretability.
完善多器官、多组学生物衰老时钟的生成、解释和应用。
来自临床表型和神经影像学数据的多器官生物衰老时钟已成为研究人类衰老和疾病的宝贵工具。血浆蛋白质组学提供了一个额外的分子维度来丰富这些时钟。在这项研究中,我开发了11个基于多器官蛋白质组的生物年龄差距(ProtBAGs),使用了来自英国生物银行43498名参与者的2448个血浆蛋白。在这里,我强调了开发和使用这些时钟的方法和临床考虑,包括纠正年龄偏差,蛋白质的器官特异性,样本量和训练数据中的潜在病理,这些都会影响模型的通用性和临床可解释性。此外,我将11个ProtBAGs与先前开发的9个基于多器官表型的生物学年龄差距相结合,以研究遗传重叠及其与疾病终点的因果关系。最后,我表明结合跨器官的特征可以提高对全身性疾病类别和全因死亡率的预测。这些分析为开发和解释这些时钟提供了方法学和临床见解,并强调了未来多器官,多组学生物衰老时钟框架的途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


