Label-Free Quantification of DNA Loading on Centrifugation-Resistant Spherical Nucleic Acids
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
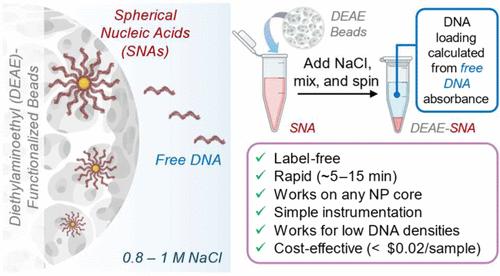
The accurate determination of DNA loading on spherical nucleic acids (SNAs) is critical for biosensing and therapeutic applications, as DNA loading directly affects target binding, cellular uptake, and therapeutic efficacy. Traditional methods involve centrifugation to separate unbound DNA from SNAs, followed by DNA quantification to calculate the number of strands per particle. However, this approach is ineffective for centrifugation-resistant SNAs. Alternative methods, such as centrifugal filtration, require multiple washing steps, often resulting in sample loss, and can be labor- and time-intensive, taking several hours. Additionally, many existing techniques rely on expensive labels or toxic reagents. Here, we present a simple, rapid, and label-free method for determining DNA loading using diethylaminoethyl (DEAE)-functionalized beads. We demonstrate its effectiveness on centrifugation-resistant SNAs, including those with metallic or protein-based cores and various DNA lengths, sequences, and densities. This approach exploits the different binding strengths of free and nanoparticle-bound DNA to DEAE, allowing the selective elution of free DNA by adjusting the ionic strength. The eluted DNA is quantified to determine the unbound fraction, which is then used to calculate the number of bound DNA strands per nanoparticle. Requiring only standard laboratory equipment─such as a benchtop centrifuge, a shaker, and either a UV–vis spectrophotometer or a commonly available plate reader─this method provides a fast and reliable alternative to conventional approaches, delivering results in under 15 min. Its versatility and broad applicability across diverse SNA core materials and sizes make it a valuable tool for DNA quantification in a wide range of nanoparticle-based platforms.
抗离心球形核酸上DNA负载的无标记定量
准确测定球形核酸(SNAs)上的DNA负载对于生物传感和治疗应用至关重要,因为DNA负载直接影响靶标结合、细胞摄取和治疗效果。传统的方法包括离心分离未结合的DNA和SNAs,然后进行DNA定量计算每个颗粒的链数。然而,这种方法对抗离心SNAs无效。替代方法,如离心过滤,需要多个洗涤步骤,经常导致样品损失,并且可能是劳动和时间密集型的,需要几个小时。此外,许多现有的技术依赖于昂贵的标签或有毒试剂。在这里,我们提出了一种简单,快速,无标记的方法来确定DNA负载使用二乙基氨基乙基(DEAE)功能化珠。我们证明了它对抗离心SNAs的有效性,包括那些具有金属或蛋白质基核心和各种DNA长度,序列和密度的SNAs。这种方法利用了游离DNA和纳米颗粒结合DNA与DEAE的不同结合强度,通过调节离子强度来选择性地洗脱游离DNA。被洗脱的DNA被量化以确定未结合的部分,然后用于计算每个纳米颗粒的结合DNA链的数量。只需要标准的实验室设备──如台式离心机、摇床、紫外可见分光光度计或常用的板读取器──这种方法提供了一种快速可靠的替代传统方法,在15分钟内提供结果。它的通用性和广泛的适用性在不同的SNA核心材料和尺寸,使其成为一个有价值的工具DNA定量在广泛的纳米颗粒为基础的平台。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


