Biallelic loss-of-function variants in ZNF142 are associated with a robust DNA methylation signature affecting a limited number of genomic loci
IF 4.6
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Biallelic inactivating variants in ZNF142 underlie a clinically variable neurodevelopmental disorder. ZNF142 is a zinc-finger transcription factor with potential roles on chromatin organization, implying a possible association of ZNF142 loss of function with perturbed genome-wide DNA methylation (DNAm) pattern. We performed EPIC array-based methylation profiling of peripheral blood-derived DNA samples from 27 individuals with biallelic ZNF142 inactivating variants, together with 6 heterozygous carriers and 40 controls. A DNAm signature discovery pipeline was applied by using 440 controls for discovery and validation analyses, and a machine-learning model was trained to classify 8 individuals carrying ZNF142 variants of uncertain clinical significance. Analyses directed to explore the genome-wide DNAm landscape in affected individuals revealed 88 differentially methylated probes constituting the minimal informative set specific to ZNF142 loss of function. This reproducible pattern of DNAm changes involved regulatory regions of a small number of genes. The DNAm signature derived from peripheral blood allowed us to diagnose individuals carrying biallelic inactivating ZNF142 variants when applied to fibroblasts. Our findings provide evidence that biallelic loss-of-function ZNF142 variants result in a specific and robust DNAm signature. The identified DNAm pattern suggests occurrence of a methylation disturbance involving a small number of loci that appears to be shared by different cell lineages.
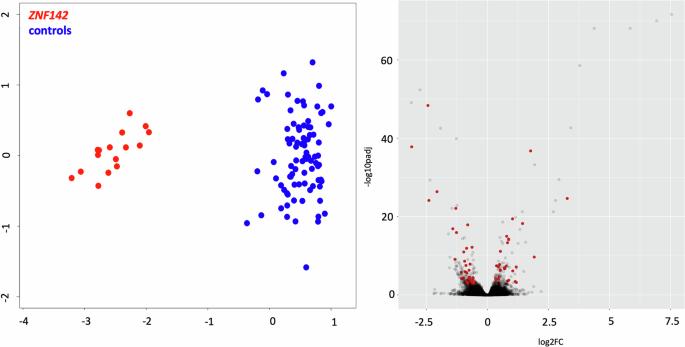
ZNF142的双等位基因功能丧失变异与影响有限数量基因组位点的强大DNA甲基化特征相关。
ZNF142的双等位基因失活变异是临床可变神经发育障碍的基础。ZNF142是一种锌指转录因子,在染色质组织中具有潜在的作用,这意味着ZNF142的功能丧失可能与全基因组DNA甲基化(DNAm)模式的紊乱有关。我们对27名ZNF142双等位基因失活变异个体、6名杂合携带者和40名对照者的外周血来源DNA样本进行了基于EPIC阵列的甲基化分析。通过使用440个对照,应用DNAm签名发现管道进行发现和验证分析,并训练机器学习模型对8个携带不确定临床意义的ZNF142变体的个体进行分类。旨在探索受影响个体全基因组dna图谱的分析显示,88个差异甲基化探针构成了ZNF142功能丧失的最小信息集。这种可重复的dna变化模式涉及少数基因的调控区域。当应用于成纤维细胞时,来自外周血的dna特征使我们能够诊断携带双等位基因失活ZNF142变体的个体。我们的研究结果提供了证据,证明双等位基因功能丧失的ZNF142变体导致了特定的和强大的dna特征。鉴定出的dna模式表明发生了涉及少数位点的甲基化干扰,这些位点似乎是由不同的细胞系共享的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

European Journal of Human Genetics
生物-生化与分子生物学
CiteScore
9.90
自引率
5.80%
发文量
216
审稿时长
2 months
期刊介绍:
The European Journal of Human Genetics is the official journal of the European Society of Human Genetics, publishing high-quality, original research papers, short reports and reviews in the rapidly expanding field of human genetics and genomics. It covers molecular, clinical and cytogenetics, interfacing between advanced biomedical research and the clinician, and bridging the great diversity of facilities, resources and viewpoints in the genetics community.
Key areas include:
-Monogenic and multifactorial disorders
-Development and malformation
-Hereditary cancer
-Medical Genomics
-Gene mapping and functional studies
-Genotype-phenotype correlations
-Genetic variation and genome diversity
-Statistical and computational genetics
-Bioinformatics
-Advances in diagnostics
-Therapy and prevention
-Animal models
-Genetic services
-Community genetics
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


