Delineation of Subcellular Molecular Heterogeneity in Single Cells via Ultralow-Flow-Rate Desorption Electrospray Ionization Mass Spectrometry Imaging
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Single-cell analysis uncovers cellular heterogeneity and dynamic metabolism at the individual cell level, providing fundamental insights into the physiological and pathological mechanisms of life. Spatial molecular mapping of single cells requires advanced techniques that achieve high spatial resolution and comprehensive molecular coverage while preserving the cells’ native state with minimal preparation. Here, we report an ultralow-flow-rate desorption electrospray ionization mass spectrometry imaging (u-DESI-MSI) platform for in situ biomolecule mapping of single cells with subcellular spatial resolution. The u-DESI-MSI single-cell platform utilizes a stable and ultralow-solvent-flow-rate (150 nL/min) system with optimized geometrical settings, avoiding complicated hardware modifications on the ESI emitter. The capability of u-DESI-MSI was demonstrated by spatial molecular mapping of human pancreatic cells from different lineages. An unprecedented spatial resolution was achieved for molecular mapping of the single cells under a rastering step size of 5 μm, even revealing lipid distribution in subcellular regions. Using this high-spatial-resolution u-DESI-MSI platform, we effectively visualized both intercellular and intracellular molecular heterogeneity in single cells. Notably, u-DESI-MSI provides a versatile tool for direct molecular imaging of single cells in their native states under ambient conditions, eliminating the need for extensive sample preparation. We anticipate that this platform will facilitate the exploration of single-cell heterogeneity, offering valuable insights into cellular variability and metabolism.
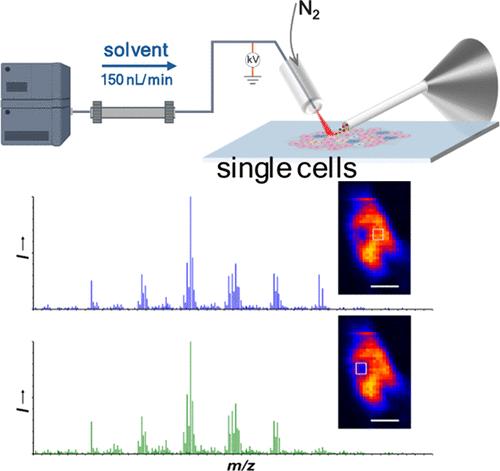
通过超低流速解吸电喷雾电离质谱成像描绘单细胞亚细胞分子异质性
单细胞分析揭示了单个细胞水平上的细胞异质性和动态代谢,为生命的生理和病理机制提供了基本的见解。单细胞的空间分子定位需要先进的技术来实现高空间分辨率和全面的分子覆盖,同时以最少的准备保留细胞的天然状态。在这里,我们报告了一个超低流速解吸电喷雾电离质谱成像(u-DESI-MSI)平台,用于亚细胞空间分辨率的单细胞原位生物分子定位。u-DESI-MSI单细胞平台采用稳定的超低溶剂流量(150 nL/min)系统,优化了几何设置,避免了ESI发射器上复杂的硬件修改。u-DESI-MSI的能力通过不同谱系的人胰腺细胞的空间分子定位得到证实。在5 μm的光栅步长下,单细胞分子作图获得了前所未有的空间分辨率,甚至揭示了亚细胞区域的脂质分布。利用这个高空间分辨率的u-DESI-MSI平台,我们有效地可视化了单个细胞中的细胞间和细胞内分子异质性。值得注意的是,u-DESI-MSI提供了一种多功能工具,可以在环境条件下对单细胞的天然状态进行直接分子成像,从而消除了大量样品制备的需要。我们预计该平台将促进单细胞异质性的探索,为细胞变异性和代谢提供有价值的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


