Deciphering the sequence-dependent unfolding pathways of an RNA pseudoknot with steered molecular dynamics
Abstract
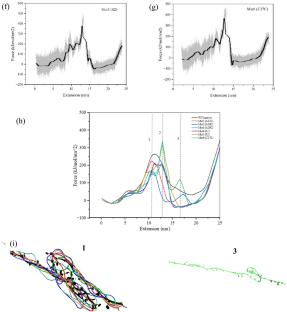
Programmed ribosomal frameshifting in Simian retrovirus-1 (SRV-1) is sensitive to the mechanical properties of an RNA pseudoknot. Unravelling these mechanical intricacies via unfolding reveals fundamental insights into their structural dynamics. Using constant velocity steered molecular dynamics (CV-SMD) simulations, we explored the unfolding dynamics and the impact of mutations on the unfolding pathway of the pseudoknot. Except for A28C, A/U to C mutations that disrupt base triples between the loop 2 and stem 1 significantly weaken the pseudoknot and make it more susceptible to unfolding. Complementary mutations in 3 base pairs of the stem region (S1) enhanced its susceptibility to disruption except for Mut5 (S2). We quantitatively assessed the variations in unfolding pathways by analysing the opening of distinct Canonical (WC) and non-canonical (NWC) interactions, force-extension curves, and potential mean force profiles (as a guiding decision for planning mutations). These findings offer a quantified perspective, showcasing the potential of utilizing the unfolding pathways of RNA pseudoknots to explore the programmability of RNA structures. This insight proves valuable for designing RNA-PROTACS and RNA-aptamers, allowing for the assessment and manipulation of their biological folding/unfolding processes.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


