Characterization and Mechanism Study of a Novel Ethanol Acetyltransferase from Hanseniaspora uvarum (EatH) with Good Thermostability, pH Stability, and Broad Alcohol Substrate Specificity
IF 5.7
1区 农林科学
Q1 AGRICULTURE, MULTIDISCIPLINARY
引用次数: 0
Abstract
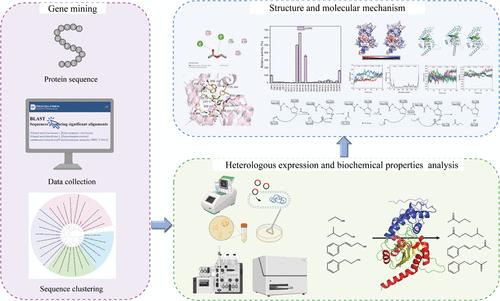
Ethyl acetate, one of the most essential industrial compounds, has a broad range of applications, including flavors, fragrances, pharmaceuticals, cosmetics, and green solvents. Eat1 is accountable for bulk ethyl acetate production in yeasts, yet its properties and molecular mechanism are not well characterized. In this study, an eat1 gene from Hanseniaspora uvarum was obtained through gene mining. EatH showed the highest activity at pH 7.5 and 35 °C and preferred short-chain acyl substrates but had a broad alcohol substrate spectrum from short-chain primary alcohols to aromatic alcohols. Its Km and kcat/Km values toward pNPA were measured to be 1.16 mM and 29.03 L·mmol–1·s–1, respectively. The structure of EatH was composed of a lid domain and a core catalytic domain, with the catalytic triad of Ser124, Asp148, and His296. Additionally, crucial residues and their mechanism were analyzed through molecular docking, site-directed mutagenesis, and molecular dynamics simulation. The mutants N149A, N149K, and N149S showed enhanced enzyme activity toward pNP-hexanoate to 5.0-, 6.6-, and 3.6-fold, and Y204S enhanced enzyme activity for pNP-butyrate by 2.6 times via creating a wider substrate binding pocket and enhancing hydrophobicity. Collectively, this work provided a theoretical basis for the further rational design of EatH and enriched the understanding of the Eat family.
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.90
自引率
8.20%
发文量
1375
审稿时长
2.3 months
期刊介绍:
The Journal of Agricultural and Food Chemistry publishes high-quality, cutting edge original research representing complete studies and research advances dealing with the chemistry and biochemistry of agriculture and food. The Journal also encourages papers with chemistry and/or biochemistry as a major component combined with biological/sensory/nutritional/toxicological evaluation related to agriculture and/or food.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


