Fluorescent Fingerprint Identification of Protein Structural Changes and Disease-Specific Amyloid Beta Aggregates Based on a Single-Nanozyme Sensor Array
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
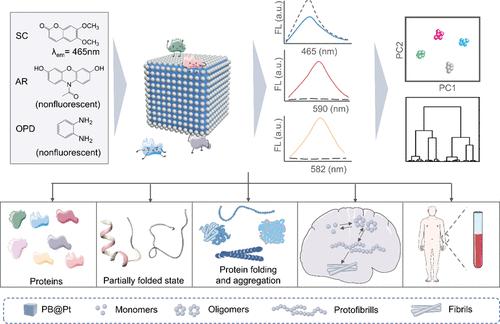
The misfolding of amyloid β (Aβ) peptides into an aggregation state is a central hallmark of the onset of Alzheimer’s disease (AD). However, conventional methods are mainly focused on detecting a specific Aβ peptide, which makes it difficult to recognize multiple analytes with different topological features and unfolded states at the same time. Here, we propose a simple and universal sensing strategy to construct a fluorescence sensor array by using a single-nanozyme probe combined with three fluorescent substrates as three recognition units to probe the protein structural changes and identify between multiple Aβ assemblies. In this sensor system, the fingerprint-like patterns are produced from the nonspecific interactions between topological proteins and the sensing units. As a result, this sensor array can accurately identify 13 kinds of proteins and their mixtures at different ratios. Moreover, the sensor array can discriminate against proteins with unfolded states and diverse conformational forms. Most importantly, the sensor array successfully distinguishes between multiple Aβ species, even in artificial cerebrospinal fluid samples and human serum samples. This work provides an attractive and reliable strategy for predicting pathologically relevant proteins and clinical diagnosis of AD.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


