Near-Infrared Light-Mediated Living Cells Mitochondrial Proteome Proximity Labeling
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
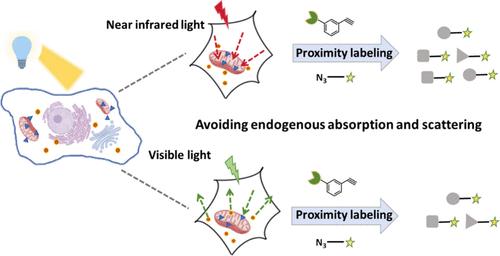
Photocatalytic proximity labeling techniques enable significant advances in understanding the subcellular proteome. However, current methods primarily utilize visible light that suffers from low labeling efficiency and limited identification coverage due to absorption overlap with endogenous chromophores and scattering in biological samples. To address these issues, we reported the proximity labeling strategy using near-infrared excitation (PL-NIR) strategy, a proximity labeling platform that used a near-infrared-excited catalyst to activate labeling mediated by reactive oxygen species, which could avoid the disadvantages of visible light. Taking advantage of the near-infrared excitation and mitochondrial targeting properties of IR780, the mitochondrial proteome were selectively labeled by spatially limited reaction in the native environment. The PL-NIR strategy facilitated the plotting of the mitochondrial proteome in which up to 245 mitochondrial proteins were identified in living HeLa cells. Compared with the current methods, the PL-NIR strategy significantly increased identification coverage, especially for mitochondrial inner membrane proteins. Furthermore, mitochondrial proteome dynamics were deciphered in LPS stimulated HMC3 cells, which were hard to transfect. Overall, the PL-NIR strategy as a highly precise proteomic platform offered improved identification coverage for more knowledge of subcellular biology discovering.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


