Ag+-Mediated DNA Nanomachine Cascade Nanomaterial Amplification Enable One-Pot Electrochemical Analysis of Circulating Tumor DNA
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
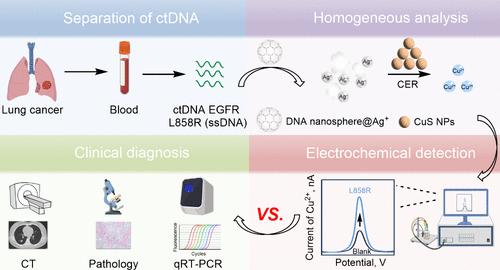
Circular tumor DNA (ctDNA) is a trace nucleic acid that functions as an essential tumor marker. In this context, the present study proposes a one-pot electrochemical analysis of ctDNA EGFR L858R in lung cancer leveraging a Ag+-mediated DNA nanosphere (I amplification) and cation exchange reaction (II amplification), and Cu2+ acts as a signal molecule. Once the target L858R exists, it specifically destroys the structure of DNA nanosphere@Ag+, and large amounts of Ag+ are released. After the addition of copper sulfide nanoparticles, Cu2+ can be replaced by a cation exchange reaction. Eventually, the electrochemical signal of Cu2+ is elevated. The analytical performance of the method is satisfactory, L858R can be detected in the linear range of 1 aM-1 fM with a detection limit of 0.3 aM. Furthermore, the system exhibits notable selectivity in differentiating base mismatch targets and other ctDNA sequences. The recovery rate of blood samples is between 95.5 and 105%. The electrochemical results from the analysis of 42 clinical blood samples are consistent with those of the quantitative real-time polymerase chain reaction, computed tomography, and pathology results. In summary, this novel strategy utilizes preprepared functional nucleic acid nanomaterials and cascade amplification, which is expected to contribute to the sensitive and expeditious detection of trace nucleic acids.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


