Molecular Profiling of Glioblastoma Patient-Derived Single Cells Using Combined MALDI-MSI and MALDI-IHC
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
In recent years, mass spectrometry-based imaging techniques have improved at unprecedented speeds, particularly in spatial resolution, and matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) experiments can now routinely image molecular profiles of single cells in an untargeted fashion. With the introduction of MALDI-immunohistochemistry (IHC), multiplexed visualization of targeted proteins in their native tissue location has become accessible and joins the suite of multimodal imaging techniques that help unravel molecular complexities. However, MALDI-IHC has not been validated for use with cell cultures at single-cell level. Here, we introduce a workflow for combining MALDI-MSI and MALDI-IHC on single, isolated cells. Patient-derived cells from glioblastoma tumor samples were imaged, first with high-resolution MSI to obtain a lipid profile, followed by MALDI-IHC highlighting cell-specific protein markers. The multimodal imaging revealed cell type specific lipid profiles when comparing glioblastoma cells and neuronal cells. Furthermore, the initial MSI measurement and its sample preparation showed no significant differences in the subsequent MALDI-IHC ion intensities. Finally, an automated recognition model was created based on the MALDI-MSI data and was able to accurately classify cells into their respective cell type in agreement with the MALDI-IHC markers, with triglycerides, phosphatidylcholines, and sphingomyelins being the most important classifiers. These results show how MALDI-IHC can provide additional valuable molecular information on single-cell measurements, even after an initial MSI measurement without reduced efficacy. Investigation of heterogeneous single-cell samples has the potential of giving a unique insight into the dynamics of how cell-to-cell interaction drives intratumor heterogeneity, thus highlighting the perspective of this work.
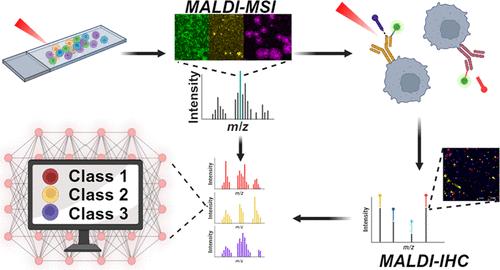
使用MALDI-MSI和MALDI-IHC联合分析胶质母细胞瘤患者来源的单细胞的分子谱
近年来,基于质谱的成像技术以前所未有的速度发展,特别是在空间分辨率方面,基质辅助激光解吸/电离(MALDI)质谱成像(MSI)实验现在可以常规地以非靶向方式成像单细胞的分子谱。随着maldi免疫组织化学(IHC)的引入,靶向蛋白在其天然组织位置的多模态可视化已经成为可能,并加入了有助于解开分子复杂性的多模态成像技术。然而,MALDI-IHC尚未被证实用于单细胞水平的细胞培养。在这里,我们介绍了在单个分离细胞上结合MALDI-MSI和MALDI-IHC的工作流程。对来自胶质母细胞瘤肿瘤样本的患者来源的细胞进行成像,首先使用高分辨率MSI获得脂质谱,然后使用MALDI-IHC突出细胞特异性蛋白质标记。当比较胶质母细胞瘤细胞和神经元细胞时,多模态成像显示细胞类型特异性脂质谱。此外,初始MSI测量及其样品制备在随后的MALDI-IHC离子强度中没有显着差异。最后,基于MALDI-MSI数据创建了一个自动识别模型,能够根据MALDI-IHC标记准确地将细胞分类为各自的细胞类型,其中甘油三酯、磷脂酰胆碱和鞘磷脂是最重要的分类器。这些结果表明MALDI-IHC可以在单细胞测量中提供额外的有价值的分子信息,即使在初始MSI测量之后也不会降低效率。异质性单细胞样本的研究有可能为细胞间相互作用如何驱动肿瘤内异质性的动力学提供独特的见解,从而突出这项工作的观点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


