Dissecting the Genetic Basis of Preharvest Sprouting in Rice Using a Genome-Wide Association Study
IF 5.7
1区 农林科学
Q1 AGRICULTURE, MULTIDISCIPLINARY
引用次数: 0
Abstract
Preharvest sprouting (PHS) is an unfavorable trait in cereal crops that significantly reduces grain yield and quality. However, the regulatory mechanisms underlying this complex trait are still largely unknown. Here, 276 rice accessions from the 3000 Rice Genomes Project were used to perform a genome-wide association study. In total, seven PHS-associated quantitative trait loci (QTLs) were identified, including two novel QTLs and five previously reported QTLs. Among them, four QTLs were identified in 2020 and 2021, and rice accessions carrying at least two favorable alleles exhibited significantly improved PHS resistance. Within these four stable QTLs, five candidate genes were identified based on haplotype and gene expression analyses, including two genes in qPhs-1.1, one gene in qPhs-3, one gene in qPhs-6, and one gene in qPhs-7. Notably, the novel QTL qPhs-6 was found to contain the candidate gene Pi starvation-induced transcription factor 1 (OsPTF1). We discovered that OsPTF1 plays a novel role in negatively regulating PHS in rice and identified two elite haplotypes of OsPTF1 associated with low PHS. Our results provide future insight into the genetic basis of PHS and will prove useful in the future studies on the role of OsPTF1 in PHS in rice.
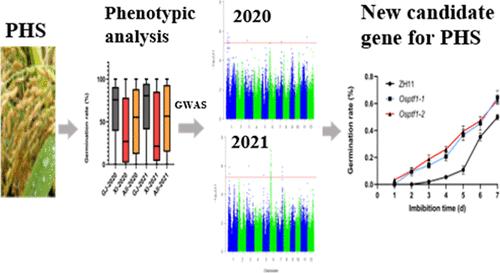
利用全基因组关联研究剖析水稻收获前发芽的遗传基础
收获前发芽是谷类作物的一个不利性状,严重影响粮食产量和品质。然而,这种复杂特征背后的调控机制在很大程度上仍然是未知的。本文利用来自3000个水稻基因组计划的276个水稻材料进行全基因组关联研究。共鉴定出7个phs相关的数量性状位点(qtl),包括2个新的qtl和5个先前报道的qtl。其中,在2020年和2021年鉴定出4个qtl,携带至少两个有利等位基因的水稻材料对小灵通的抗性显著提高。在这4个稳定的QTLs中,通过单倍型和基因表达分析鉴定出5个候选基因,其中2个基因位于qPhs-1.1, 1个基因位于qPhs-3, 1个基因位于qPhs-6, 1个基因位于qPhs-7。值得注意的是,新的QTL qPhs-6被发现含有候选基因Pi饥饿诱导转录因子1 (OsPTF1)。我们发现OsPTF1在水稻小灵通负调控中发挥了新的作用,并鉴定出两个与低小灵通相关的OsPTF1精英单倍型。我们的研究结果为小灵通的遗传基础提供了进一步的见解,并将为未来研究OsPTF1在水稻小灵通中的作用提供帮助。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.90
自引率
8.20%
发文量
1375
审稿时长
2.3 months
期刊介绍:
The Journal of Agricultural and Food Chemistry publishes high-quality, cutting edge original research representing complete studies and research advances dealing with the chemistry and biochemistry of agriculture and food. The Journal also encourages papers with chemistry and/or biochemistry as a major component combined with biological/sensory/nutritional/toxicological evaluation related to agriculture and/or food.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


