Localized Bicirculating DNAzyme Self-Feedback Amplification Strategy for Ultra-Sensitive Fluorescence Biosensing of MicroRNA
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
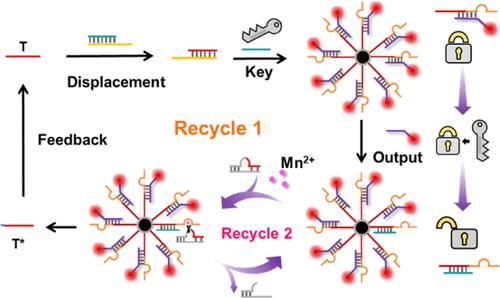
DNAzyme-based cascade networks are effective tools to achieve ultrasensitive detection of low-abundance miRNAs. However, their designs are complicated and costly, and the operation is time-consuming. Herein, a novel simple noncascade DNAzyme network is designed and its amplification effect is comparable to or even better than many cascading ones. It is a nonenzymatic, isothermal, bicirculating amplification network consisting of two toehold-mediated strand-displacement reactions and a localized DNAzyme amplification strategy. Taking microRNA-122 as a target model, this ultrasensitive fluorescence biosensor has a detection limit of 84 zmol L–1, which is 8-orders of magnitude lower than that of the nonamplification one. The ultrasensitivity mainly benefits from the exclusive design and positive self-feedback mechanism of the ingenious bicirculating DNAzyme amplification network. In addition, the utilization of superparamagnetic Fe3O4@SiO2 particles not only helps for the localization of DNAzymes but also facilitates the rapid separation of signal probes (output DNA-CdTe QDs). This fluorescent biosensor also has the advantages of specificity, speed, thermal stability, and low cost. This novel design paves a new way to simple and effective bioamplification strategy, which may be very attractive for biosensors, DNA logic gates, and DNA computers.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


