Accurate Classification of Human CD4+ T, CD8+ T, and CD19+ B Cells Isolated from Splenocytes by Cross-Polarized Diffraction Image Pairs
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Diffraction imaging of cells allows rapid phenotyping by the response of intracellular molecules to coherent illumination. However, its ability to distinguish numerous types of human leukocytes remains to be investigated. Here, we show that accurate classification of three lymphocyte subtypes can be achieved with features extracted from cross-polarized diffraction image (p-DI) pairs. A deep neural network (DNN) of DINet-PS has been developed for feature extraction from and filtering of, in the angular frequency domain, p-DI pairs acquired from live lymphocytes isolated from human spleen tissues. We built the network in a dual-channel structure and incorporated two adaptive spectral filter blocks to actively suppress extracted features related to the noise component of light in p-DI pairs. The DINet-PS was trained with p-DI pairs acquired from 5311 CD4+ T, 3819 CD8+ T, and 4054 CD19+ B cells after preprocessing and rebelling of manually derived secondary labels and classification accuracy of 96.6 ± 0.40% has been achieved in hold-out test data sets among the three subtypes. Our results show the power of DNN to extract cell-related features from p-DI pairs and the potential of polarization diffraction imaging flow cytometry for accurate and label-free classification of lymphocyte subtypes in particular and leukocytes in general.
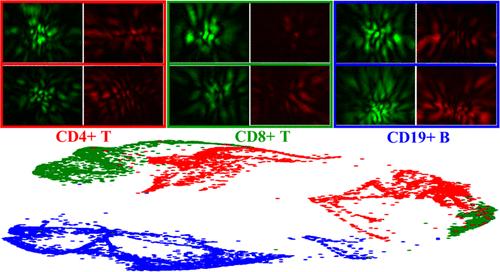
交叉偏振衍射图像对对人脾细胞CD4+ T、CD8+ T和CD19+ B细胞的准确分类
细胞的衍射成像允许通过细胞内分子对相干照明的响应快速表型。然而,其区分多种人类白细胞的能力仍有待研究。在这里,我们表明可以通过交叉偏振衍射图像(p-DI)对提取的特征来实现三种淋巴细胞亚型的准确分类。本文开发了一种基于DINet-PS的深度神经网络(DNN),用于从人脾组织中分离的活淋巴细胞中提取p-DI对的角频域特征提取和滤波。我们构建了双通道结构的网络,并加入了两个自适应光谱滤波块,以主动抑制p-DI对中与光噪声成分相关的提取特征。DINet-PS采用5311个CD4+ T、3819个CD8+ T和4054个CD19+ B细胞的p-DI对进行预处理和人工生成二级标签后进行训练,在三种亚型的保持测试数据集中,分类准确率达到96.6±0.40%。我们的研究结果显示了DNN从p-DI对中提取细胞相关特征的能力,以及偏振衍射成像流式细胞术在准确和无标记分类淋巴细胞亚型和一般白细胞方面的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


