Signal Processing Workflow for Suspect Screening in LC × LC-HRMS: Efficient Extraction of Pure Mass Spectra for Identification of Suspects in Complex Samples Using a Mass Filtering Algorithm
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
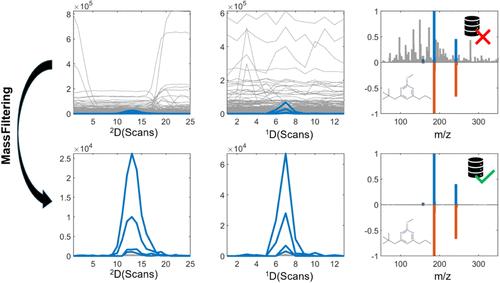
The data processing workflows for comprehensive two-dimensional liquid chromatography (LC × LC) hyphenated to high-resolution mass spectrometry (HRMS) operated in data-independent acquisition (DIA) are limited compared to their one-dimensional counterparts. A two-step workflow is proposed to extract pure mass spectra from LC × LC-HRMS. First, a mass filtering (MF) algorithm groups ions belonging to the same compound based on their elution profile similarity in the first (1D) and second dimension (2D). Second, the filtered data are deconvoluted using multivariate curve resolution (MCR) to address potential coelution. The presented workflow is termed MF + MCR and was tested on pulsed elution-LC × LC-HRMS data from a wastewater effluent extract. The proposed workflow was benchmarked to the following three data processing strategies for mass spectra extraction: peak apex (PAM), using the MF approach alone, or using MCR without prior MF. The MF + MCR workflow identified 25 suspect compounds, compared to 23, 16, and 10 identified by MF, MCR, and PAM, respectively. The nine suspects that could not be identified using MCR compared to the MF + MCR all had low total signal contributions, i.e., low intensities compared to the TIC. This showed that adequate preprocessing prior to MCR is essential for trace level analysis. Additionally, it was shown that the MF + MCR workflow extracted statistically significantly purer mass spectra compared to PAM (p-value: 0.003) and MCR (p-value: 0.04) from a spiked blank sample. The results highlight that by utilizing the elution profiles in both chromatographic dimensions, clean mass spectra of analytes at trace levels measured in DIA can be extracted, allowing for more reliable compound identification compared to the workflows that were used for benchmarking.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


