Millisecond Label-Free Single Peptide Detection and Identification Using Nanoscale Electrochromatography and Resistive Pulse Sensing
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
We are developing a unique protein identification method that consists of generating peptides proteolytically from a single protein molecule (i.e., peptide fingerprints) with peptide detection and identification carried out using nanoscale electrochromatography and label-free resistive pulse sensing (RPS). As a step in realizing this technology, we report herein the nanoscale electrochromatography of model peptides using thermoplastic columns with surfaces engineered to identify peptides via their molecularly dependent mobility (i.e., time-of-flight, ToF). ToFs were elucidated using a dual in-plane nanopore sensor, which consisted of two in-plane nanopores placed on either end of the nanoelectrochromatography column. The surface of the nanocolumn, which consisted of poly(methyl methacrylate) (PMMA), was activated with an O2 plasma, creating surface carboxylic acid groups (−COOH) inducing a surface charge on the column wall as well as affecting its hydrophilicity. To understand scaling effects, we carried out microchip and nanochannel electrochromatography of the peptides labeled with an ATTO 532 reporter to allow for single-molecule tracking. Our results indicated that the apparent mobilities of the model peptides did not allow for their separation in a microchannel, but when performed in a nanocolumn, clear differences in their apparent mobilities could be observed especially when operated at high electric field strengths. We next performed label-free detection of peptides using the dual in-plane nanopore sensor with the two pores separated by a 5 μm (length) column with a 50 nm width and depth. When a single peptide molecule passed through an in-plane nanopore, the sensor read a pair of resistive pulses with a time difference equivalent to ToF. We identified the peptides by evaluating their ToF, normalized RPS current transient amplitude (ΔI/I0), and RPS peak dwell time (td). We could identify the model peptides with nearly 100% classification accuracy at the single-molecule level using machine learning with a single molecule measurement requiring <10 ms.
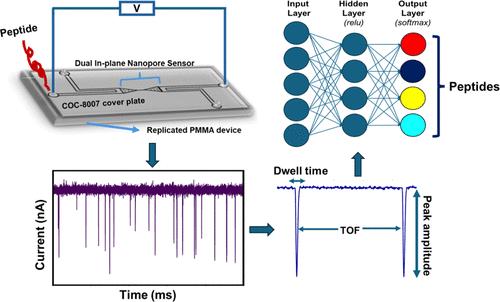
利用纳米电色谱和电阻脉冲传感技术检测和鉴定毫秒无标记单肽
我们正在开发一种独特的蛋白质鉴定方法,该方法包括从单个蛋白质分子(即肽指纹)中水解生成肽,并使用纳米级电色谱和无标签电阻脉冲传感(RPS)进行肽检测和鉴定。作为实现这项技术的一步,我们在此报告了模型肽的纳米级电色谱,使用热塑性柱,表面经过设计,通过分子依赖的迁移率(即飞行时间,ToF)来识别肽。利用双平面内纳米孔传感器,在纳米电色谱柱的两端分别放置两个平面内纳米孔,对tfs进行了表征。由聚甲基丙烯酸甲酯(PMMA)组成的纳米柱的表面被O2等离子体活化,产生表面羧酸基团(- COOH),在柱壁上诱导表面电荷,并影响其亲水性。为了理解标度效应,我们对ATTO 532报告基因标记的肽进行了微芯片和纳米通道电色谱分析,以实现单分子跟踪。我们的研究结果表明,模型肽的表观流动性不允许它们在微通道中分离,但当在纳米柱中进行时,它们的表观流动性可以观察到明显的差异,特别是在高电场强度下操作时。接下来,我们使用双平面内纳米孔传感器进行免标记检测,两个孔由5 μm(长)的柱隔开,宽度和深度为50 nm。当单个肽分子通过平面内纳米孔时,传感器读取一对时间差相当于ToF的电阻脉冲。我们通过评估它们的ToF、归一化RPS电流瞬态振幅(ΔI/I0)和RPS峰停留时间(td)来鉴定肽。我们可以使用机器学习在单分子水平上以接近100%的分类准确率识别模型肽,单分子测量需要<;10 ms。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


