Personalized Profiling of Lipoprotein and Lipid Metabolism Based on 1018 Measures from Combined Quantitative NMR and LC-MS/MS Platforms
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Applications of advanced omics methodologies are increasingly popular in biomedicine. However, large-scale studies aiming at clinical translation are typically siloed to single technologies. Here, we present the first comprehensive large-scale population data combining 209 lipoprotein measures from a quantitative NMR spectroscopy platform and 809 lipid classes and species from a quantitative LC-MS/MS platform. These data with 1018 molecular measures were analyzed in two population cohorts totaling 7830 participants. The association and cluster analyses revealed excellent coherence between the methodologically independent data domains and confirmed their quantitative compatibility and suitability for large-scale studies. The analyses elucidated the detailed molecular characteristics of the heterogeneous circulatory macromolecular lipid transport system and the underlying structural and compositional relationships. Unsupervised neural network analysis─the so-called self-organizing maps (SOMs)─revealed that these deep molecular and metabolic data are inherently related to key physiological and clinical population characteristics. The data-driven population subgroups uncovered marked differences in the population distribution of multiple cardiometabolic risk factors. These include, e.g., multiple lipoprotein lipids, apolipoprotein B, ceramides, and oxidized lipids. All 79 structurally unique triglyceride species showed similar associations over the entire lipoprotein cascade and indicated systematically increased risk for carotid intima media thickening and other atherosclerosis risk factors, including obesity and inflammation. The metabolic attributes for 27 individual cholesteryl ester species, which formed six distinct clusters, were more intricate with associations both with higher─e.g., CE(16:1)─and lower─e.g., CE(20:4)─cardiometabolic risk. The molecular details provided by these combined data are unprecedented for molecular epidemiology and demonstrate a new potential avenue for population studies.
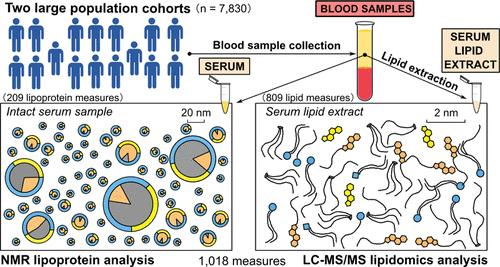
基于1018个定量NMR和LC-MS/MS平台的脂蛋白和脂质代谢个性化分析
在生物医学领域,先进的全息方法的应用日益普及。然而,以临床转化为目标的大规模研究通常都是孤立的单一技术。在这里,我们首次展示了全面的大规模人群数据,这些数据结合了来自定量核磁共振光谱平台的 209 种脂蛋白测量方法和来自定量 LC-MS/MS 平台的 809 种脂质类别和种类。这些数据包含 1018 项分子指标,对两个人群队列共计 7830 名参与者进行了分析。关联分析和聚类分析显示,这两个在方法上独立的数据域之间具有极好的一致性,并证实了它们在定量方面的兼容性和对大规模研究的适用性。这些分析阐明了异质循环大分子脂质运输系统的详细分子特征以及潜在的结构和组成关系。无监督神经网络分析--即所谓的自组织图(SOM)--揭示了这些深层分子和代谢数据与关键的生理和临床人群特征之间的内在联系。数据驱动的人群分组揭示了多种心脏代谢风险因素在人群中分布的明显差异。这些因素包括多种脂蛋白脂质、载脂蛋白 B、神经酰胺和氧化脂质等。所有 79 种结构独特的甘油三酯在整个脂蛋白级联中显示出相似的关联,并表明颈动脉内膜增厚和其他动脉粥样硬化风险因素(包括肥胖和炎症)的风险系统性增加。27 种胆固醇酯的代谢属性形成了六个不同的群组,它们与较高的(如 CE(16:1))和较低的(如 CE(20:4))心血管代谢风险都有关联。这些综合数据所提供的分子细节对于分子流行病学来说是史无前例的,为人群研究提供了一条新的潜在途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


