Data-Independent Acquisition–Parallel Reaction Monitoring Acquisition Reveals Age-Dependent Alterations of the Lysosomal Proteome in a Mouse Model of Metachromatic Leukodystrophy
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
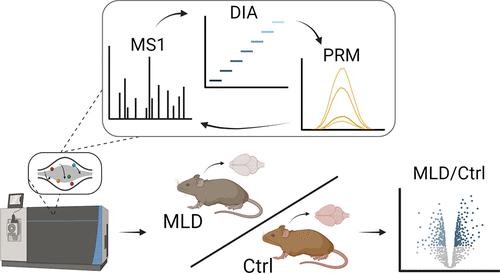
For the reproducible analysis of peptides by mass spectrometry-based proteomics, data-independent acquisition (DIA) and parallel/multiple reaction monitoring (PRM/MRM) deliver unrivalled performance with respect to sensitivity and reproducibility. Both approaches, however, come with distinct advantages and shortcomings. While DIA enables unbiased whole proteome analysis, it shows limitations with respect to dynamic range and the quantification of low-abundant proteins. PRM, on the other hand, is ideally suited to reproducibly quantify selected proteins even if they are low-abundant, but no knowledge of the remaining sample is obtained. Here, we combine both methods into a mixed DIA-PRM acquisition approach, merging their benefits while operating at reduced machine run times and needed sample amounts. We demonstrate the feasibility of DIA-PRM by merging a scheduled PRM assay for 103 peptides, representing 59 low-abundant lysosomal hydrolases, with a DIA data acquisition scheme. After benchmarking DIA-PRM with mouse embryonic fibroblast (MEF) whole cell lysates, we use the approach to investigate age-related proteomic changes in brain tissues of a mouse model of metachromatic leukodystrophy (MLD). This revealed an MLD-related progressive increase in distinct classes of lysosomal hydrolases as well as alterations of proteins related to myelin and cellular metabolism. All data are available via ProteomeXchange with PXD052313.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


