TAggiXL: A Fluorescence-Traceable Cross-Linking Strategy for Unbiased Profiling of Protein Aggregation and Interactome Dynamics
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Protein aggregation is a hallmark of numerous degenerative diseases, yet its underlying mechanisms remain poorly understood due to the challenges in identifying the composition and interaction networks of these aggregates. To address this issue, we developed TAggiXL, a novel method that combines fluorescence-traceable aggregate isolation with cross-linking proteomics, significantly enhancing the efficiency and precision of isolating protein aggregates. This method facilitates unbiased profiling of aggregated proteomes and their interactomes in live cells. The TAggiXL approach leverages advanced cross-linking proteomics, density gradient centrifugation, and fluorescence tracking to provide detailed characterization of protein aggregation under various stress conditions including HSP90 and proteasome inhibition. Using TAggiXL, we identified key components and interactions within the aggregates, particularly highlighting E3 ubiquitin ligase TRIM26, which plays a crucial role in aggregate formation and autophagic clearance under stress and pathogenic conditions. Moreover, TAggiXL revealed that HSPA1B functions as a central interaction hub within the aggregated proteome. It preferentially interacts with intrinsically disordered regions (IDRs) of aggregate components and demonstrates dynamic behavior within the aggregate. In summary, TAggiXL offers a powerful tool for dissecting the complex composition and interaction networks of protein aggregates, with a significant potential to advance our understanding of protein aggregation in degenerative diseases. It also holds promise for the development of future therapeutic interventions.
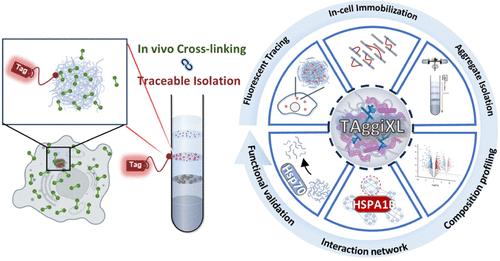
TAggiXL:用于蛋白质聚集和相互作用组动态无偏分析的荧光可追踪交联策略
蛋白质聚集是许多退行性疾病的标志,但由于难以确定这些聚集体的组成和相互作用网络,人们对其潜在机制仍然知之甚少。为了解决这个问题,我们开发了 TAggiXL,这是一种将荧光可追踪聚集体分离与交联蛋白质组学相结合的新方法,大大提高了分离蛋白质聚集体的效率和精度。这种方法有助于对活细胞中的聚集蛋白质组及其相互作用组进行无偏分析。TAggiXL方法利用先进的交联蛋白质组学、密度梯度离心和荧光追踪技术,详细描述了在各种应激条件(包括HSP90和蛋白酶体抑制)下蛋白质聚集的特征。利用TAggiXL,我们确定了聚集体中的关键成分和相互作用,尤其突出了E3泛素连接酶TRIM26,它在应激和致病条件下的聚集体形成和自噬清除中发挥着关键作用。此外,TAggiXL 还揭示了 HSPA1B 在聚集蛋白质组中的中心交互枢纽作用。它优先与聚集体成分的内在无序区(IDR)相互作用,并在聚集体中表现出动态行为。总之,TAggiXL 为剖析蛋白质聚集体的复杂组成和相互作用网络提供了一个强大的工具,极有可能促进我们对退行性疾病中蛋白质聚集的了解。它还为开发未来的治疗干预措施带来了希望。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


