Design and Validation of Specific Oligonucleotide Probes on Planar Magnetic Biosensors
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
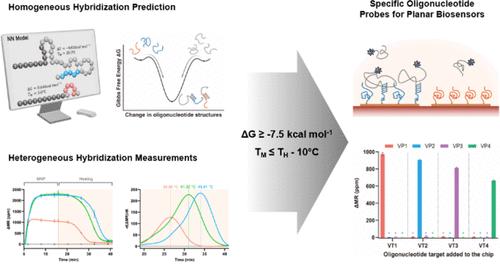
Planar DNA biosensors employ surface-tethered oligonucleotide probes to capture target molecules for diagnostic applications. To improve the sensitivity and specificity of biosensing, hybridization affinities should be enhanced, and cross-hybridization with off-targets must be minimized. To this end, assays can be designed using the thermodynamic properties of hybridization between probes and on-targets or off-targets based on Gibbs free energies and melting temperatures. However, the nature of heterogeneous hybridization between the probes on the surface and the targets in a solution imposes challenges in predicting precise hybridization affinities and the degree of cross-hybridization due to indeterminable thermodynamic penalties induced by the solid surface and its status. Herein, we suggest practical and convenient guidelines for designing oligonucleotide probes based on data obtained from planar magnetic biosensors and thermodynamic properties calculated by using easily accessible solution-phase prediction. The suggested requirements comprised Gibbs free energy ≥ −7.5 kcal mol–1 and melting temperature ≤10 °C below the hybridization temperature, and we validated for the absence of cross-hybridization. Additionally, the effects of secondary structures such as hairpins and homodimers were investigated for better oligonucleotide probe designs. We believe that these practical guidelines will assist researchers in developing planar magnetic biosensors with high sensitivity and specificity for the detection of new targets.
平面磁性生物传感器上特异性寡核苷酸探针的设计与验证
平面 DNA 生物传感器采用表面系留寡核苷酸探针捕获目标分子,用于诊断应用。为提高生物传感的灵敏度和特异性,应增强杂交亲和力,并尽量减少与非目标的交叉杂交。为此,可以根据吉布斯自由能和熔化温度,利用探针与靶上或靶下杂交的热力学特性来设计检测方法。然而,表面探针与溶液中靶标之间的异质杂交性质给预测精确的杂交亲和力和交叉杂交程度带来了挑战,因为固体表面及其状态引起的热力学惩罚是不确定的。在此,我们根据从平面磁性生物传感器获得的数据和利用易于获得的溶液相预测计算出的热力学性质,提出了设计寡核苷酸探针的实用便捷指南。建议的要求包括吉布斯自由能≥ -7.5 kcal mol-1,熔化温度低于杂交温度≤10 °C,我们还验证了无交叉杂交。此外,我们还研究了二级结构(如发夹和同源二聚体)对更好的寡核苷酸探针设计的影响。我们相信,这些实用指南将有助于研究人员开发具有高灵敏度和特异性的平面磁性生物传感器,用于检测新的目标。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


