Barley Grain Proteome Assessment Using Multi-Environment Trial Data and Machine Learning
IF 6.2
1区 农林科学
Q1 AGRICULTURE, MULTIDISCIPLINARY
引用次数: 0
Abstract
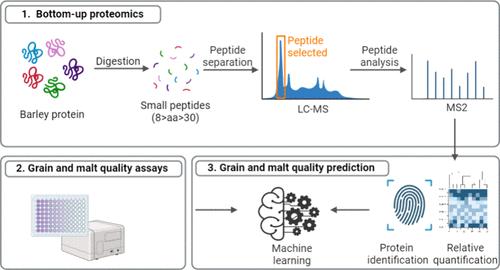
Proteomics can be used to assess individual protein abundances, which could reflect genotypic and environmental effects and potentially predict grain/malt quality. In this study, 79 barley grain samples (genotype-location-year combinations) from Californian multi-environment trials (2017–2022) were assessed using liquid chromatography–mass spectrometry. In total, 3104 proteins were identified across all of the samples. Location, genotype, and year explained 26.7, 17.1, and 14.3% of the variance in the relative abundance of individual proteins, respectively. Sixteen proteins with storage, DNA/RNA binding, or enzymatic functions were significantly higher/lower in abundance (compared to the overall mean) in the Yolo 3 and Imperial Valley locations, Butta 12 and LCS Odyssey genotypes, and the 2017–18 and 2021–22 years. Individual protein abundances were reasonably predictive (RMSECV = 1.25–2.04%) for total, alcohol-soluble, and malt protein content and malt fine extract. This study illustrates the role of the environment in the barley proteome and the utility of proteomics and machine learning to predict grain/malt quality.
利用多环境试验数据和机器学习评估大麦谷物蛋白质组
蛋白质组学可用于评估单个蛋白质丰度,从而反映基因型和环境影响,并有可能预测谷物/麦芽质量。在本研究中,我们使用液相色谱-质谱联用技术对加利福尼亚多环境试验(2017-2022 年)中的 79 个大麦谷物样本(基因型-地点-年份组合)进行了评估。所有样本中共鉴定出 3104 种蛋白质。地点、基因型和年份分别解释了单个蛋白质相对丰度差异的 26.7%、17.1% 和 14.3%。在 Yolo 3 和帝王谷地点、Butta 12 和 LCS Odyssey 基因型以及 2017-18 年和 2021-22 年,16 种具有储存、DNA/RNA 结合或酶功能的蛋白质丰度明显较高/较低(与总体平均值相比)。单个蛋白质丰度对总蛋白、醇溶蛋白、麦芽蛋白含量和麦芽细提取物具有合理的预测性(RMSECV = 1.25-2.04%)。这项研究说明了环境在大麦蛋白质组中的作用,以及蛋白质组学和机器学习在预测谷物/麦芽质量方面的实用性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.90
自引率
8.20%
发文量
1375
审稿时长
2.3 months
期刊介绍:
The Journal of Agricultural and Food Chemistry publishes high-quality, cutting edge original research representing complete studies and research advances dealing with the chemistry and biochemistry of agriculture and food. The Journal also encourages papers with chemistry and/or biochemistry as a major component combined with biological/sensory/nutritional/toxicological evaluation related to agriculture and/or food.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


