CRISPR/Cas9-Mediated Genome Editing of T4 Bacteriophage for High-Throughput Antimicrobial Susceptibility Testing
IF 6.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
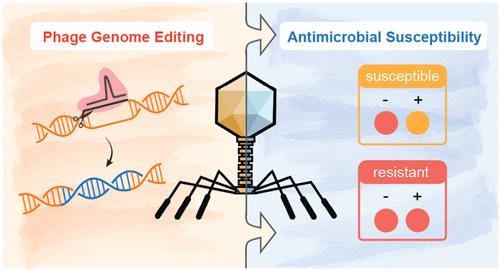
The accurate and effective determination of antimicrobial resistance is essential to limiting the spread of infectious diseases and ensuring human health. Herein, a simple, accurate, and high-throughput phage-based colorimetric sensing strategy was developed for antimicrobial susceptibility testing (AST). Taking advantage of the CRISPR/Cas9 system, the genome of the T4 phage was modularly engineered to carry lacZ-α (lacZa), a marker gene encoding the α-fragment of β-galactosidase (β-gal). T4lacZa phages were identified by blue-white selection and then used for a biosensing application. In this strategy, the bacterial solution is exposed to the T4lacZa phage, causing target bacteria to overexpress β-gal. Upon the addition of a colorimetric substrate, the β-gal initiates an enzymatic reaction, resulting in a solution color change from yellow to red. This sensing strategy offers a visual way to monitor bacterial growth in the presence of antibiotics, enabling the determination of bacterial antimicrobial susceptibility. As a proof of concept, our developed sensing strategy was successfully applied to identify 9 different multidrug-resistant Escherichia coli (E. coli) in urine samples with 100% specificity. Compared with conventional disk diffusion susceptibility tests, the engineered phage-based sensing strategy can shorten the detection time by at least half without losing detection sensitivity, providing an alternative high-throughput method for AST in clinical diagnosis.
CRISPR/Cas9 介导的 T4 噬菌体基因组编辑用于高通量抗菌药物敏感性测试
准确有效地确定抗菌素耐药性对于限制传染病的传播和确保人类健康至关重要。在此,我们开发了一种简单、准确和高通量的基于噬菌体的比色传感策略,用于抗菌药物敏感性测试(AST)。利用CRISPR/Cas9系统的优势,T4噬菌体的基因组被模块化设计为携带lacZ-α(lacZa),这是一种编码β-半乳糖苷酶(β-gal)α片段的标记基因。T4lacZa 噬菌体是通过蓝白选择鉴定出来的,然后用于生物传感应用。在这一策略中,细菌溶液暴露在 T4lacZa 噬菌体中,使目标细菌过量表达 β-gal,当加入比色底物时,β-gal 启动酶促反应,导致溶液颜色从黄色变为红色。这种传感策略提供了一种可视化的方法来监测细菌在抗生素存在下的生长情况,从而确定细菌对抗生素的敏感性。作为概念验证,我们开发的传感策略成功地用于鉴定尿液样本中 9 种不同的耐多药大肠杆菌(E. coli),特异性达到 100%。与传统的磁盘扩散药敏试验相比,基于噬菌体的工程传感策略可在不降低检测灵敏度的情况下将检测时间缩短至少一半,为临床诊断中的 AST 提供了另一种高通量方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical Chemistry
化学-分析化学
CiteScore
12.10
自引率
12.20%
发文量
1949
审稿时长
1.4 months
期刊介绍:
Analytical Chemistry, a peer-reviewed research journal, focuses on disseminating new and original knowledge across all branches of analytical chemistry. Fundamental articles may explore general principles of chemical measurement science and need not directly address existing or potential analytical methodology. They can be entirely theoretical or report experimental results. Contributions may cover various phases of analytical operations, including sampling, bioanalysis, electrochemistry, mass spectrometry, microscale and nanoscale systems, environmental analysis, separations, spectroscopy, chemical reactions and selectivity, instrumentation, imaging, surface analysis, and data processing. Papers discussing known analytical methods should present a significant, original application of the method, a notable improvement, or results on an important analyte.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


