Burden re-analysis of neurodevelopmental disorder cohorts for prioritization of candidate genes
IF 3.7
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
This study aimed to uncover novel genes associated with neurodevelopmental disorders (NDD) by leveraging recent large-scale de novo burden analysis studies to enhance a virtual gene panel used in a diagnostic setting. We re-analyzed historical trio-exome sequencing data from 745 individuals with NDD according to the most recent diagnostic standards, resulting in a cohort of 567 unsolved individuals. Next, we designed a virtual gene panel containing candidate genes from three large de novo burden analysis studies in NDD and prioritized candidate genes by stringent filtering for ultra-rare de novo variants with high pathogenicity scores. Our analysis revealed an increased burden of de novo variants in our selected candidate genes within the unsolved NDD cohort and identified qualifying de novo variants in seven candidate genes: RIF1, CAMK2D, RAB11FIP4, AGO3, PCBP2, LEO1, and VCP. Clinical data were collected from six new individuals with de novo or inherited LEO1 variants and three new individuals with de novo PCBP2 variants. Our findings add additional evidence for LEO1 as a risk gene for autism and intellectual disability. Furthermore, we prioritize PCBP2 as a candidate gene for NDD associated with motor and language delay. In summary, by leveraging de novo burden analysis studies, employing a stringent variant filtering pipeline, and engaging in targeted patient recruitment, our study contributes to the identification of novel genes implicated in NDDs.
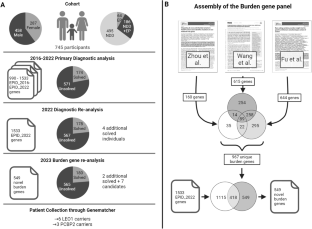
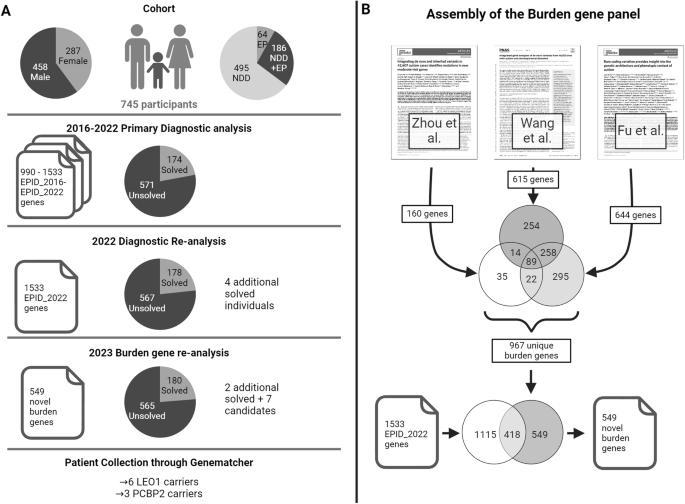
对神经发育障碍队列进行负担再分析,以确定候选基因的优先次序。
本研究旨在利用最近的大规模新基因负担分析研究,发现与神经发育障碍(NDD)相关的新基因,从而增强诊断环境中使用的虚拟基因面板。我们根据最新的诊断标准重新分析了来自 745 名 NDD 患者的历史三外显子组测序数据,得出了一个包含 567 名未决患者的队列。接下来,我们设计了一个虚拟基因面板,其中包含来自三项大型NDD从头基因负担分析研究的候选基因,并通过严格筛选具有高致病性评分的超罕见从头变异,确定候选基因的优先级。我们的分析表明,在未解决的 NDD 队列中,我们所选候选基因的从头变异负担增加,并在七个候选基因中发现了合格的从头变异:RIF1、CAMK2D、RAB11FIP4、AGO3、PCBP2、LEO1 和 VCP。我们收集了六名新的 LEO1 基因变异者或遗传性 LEO1 基因变异者和三名新的 PCBP2 基因变异者的临床数据。我们的研究结果为 LEO1 成为自闭症和智障的风险基因提供了更多证据。此外,我们还将 PCBP2 列为与运动和语言发育迟缓相关的 NDD 候选基因。总之,通过利用从头负担分析研究、采用严格的变异过滤管道和有针对性地招募患者,我们的研究有助于鉴定与 NDDs 有关的新基因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

European Journal of Human Genetics
生物-生化与分子生物学
CiteScore
9.90
自引率
5.80%
发文量
216
审稿时长
2 months
期刊介绍:
The European Journal of Human Genetics is the official journal of the European Society of Human Genetics, publishing high-quality, original research papers, short reports and reviews in the rapidly expanding field of human genetics and genomics. It covers molecular, clinical and cytogenetics, interfacing between advanced biomedical research and the clinician, and bridging the great diversity of facilities, resources and viewpoints in the genetics community.
Key areas include:
-Monogenic and multifactorial disorders
-Development and malformation
-Hereditary cancer
-Medical Genomics
-Gene mapping and functional studies
-Genotype-phenotype correlations
-Genetic variation and genome diversity
-Statistical and computational genetics
-Bioinformatics
-Advances in diagnostics
-Therapy and prevention
-Animal models
-Genetic services
-Community genetics
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


