Computational workflow for discovering small molecular binders for shallow binding sites by integrating molecular dynamics simulation, pharmacophore modeling, and machine learning: STAT3 as case study
Abstract
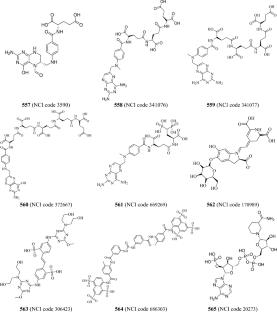
STAT3 belongs to a family of seven transcription factors. It plays an important role in activating the transcription of various genes involved in a variety of cellular processes. High levels of STAT3 are detected in several types of cancer. Hence, STAT3 inhibition is considered a promising therapeutic anti-cancer strategy. However, since STAT3 inhibitors bind to the shallow SH2 domain of the protein, it is expected that hydration water molecules play significant role in ligand-binding complicating the discovery of potent binders. To remedy this issue, we herein propose to extract pharmacophores from molecular dynamics (MD) frames of a potent co-crystallized ligand complexed within STAT3 SH2 domain. Subsequently, we employ genetic function algorithm coupled with machine learning (GFA-ML) to explore the optimal combination of MD-derived pharmacophores that can account for the variations in bioactivity among a list of inhibitors. To enhance the dataset, the training and testing lists were augmented nearly a 100-fold by considering multiple conformers of the ligands. A single significant pharmacophore emerged after 188 ns of MD simulation to represent STAT3-ligand binding. Screening the National Cancer Institute (NCI) database with this model identified one low micromolar inhibitor most likely binds to the SH2 domain of STAT3 and inhibits this pathway.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


