EasyDAM_V3:基于最优源域选择和基于知识图的数据综合的水果自动标注。
IF 6.4
1区 农林科学
Q1 AGRONOMY
引用次数: 0
摘要
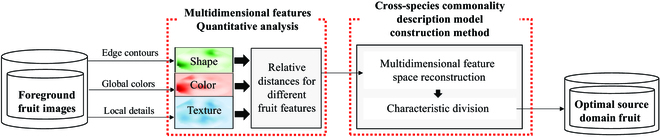
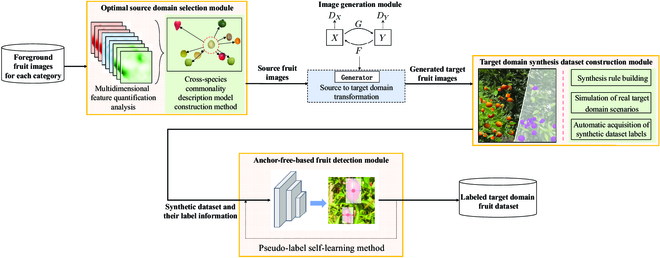
尽管基于深度学习的水果检测技术正变得越来越流行,但它们需要大量的标记数据集来支持模型训练。此外,手工贴标过程既耗时又费力。我们之前实现了一种基于生成对抗网络的方法来降低标签成本。然而,它没有考虑到更多物种之间的适应性。基于目标域水果数据集选择最合适的源域数据集的方法还有待研究。而且,目前的自动标注技术仍然需要对源领域数据集进行人工标注,不能完全消除人工过程。因此,本研究提出了一种改进的EasyDAM_V3模型,作为额外类别水果的自动标记方法。本研究提出了一种基于多维空间特征模型的最优源域建立方法,选择最合适的源域;提出了一种基于透明背景水果图像翻译的大容量数据集构建方法,构建果园场景层次成分合成规则知识图。EasyDAM_V3模型可以自动从数据集中获取水果标签信息,从而消除人工标注。以梨为优选源域,以橘子、苹果和番茄为目标域数据集,对该方法进行了验证。结果表明,目标数据集的平均标注精度分别达到90.94%、89.78%和90.84%。EasyDAM_V3模型可以在自动标注任务中获得最优的源域,从而消除了人工标注过程,降低了相关成本和人工。本文章由计算机程序翻译,如有差异,请以英文原文为准。

EasyDAM_V3: Automatic Fruit Labeling Based on Optimal Source Domain Selection and Data Synthesis via a Knowledge Graph.
Although deep learning-based fruit detection techniques are becoming popular, they require a large number of labeled datasets to support model training. Moreover, the manual labeling process is time-consuming and labor-intensive. We previously implemented a generative adversarial network-based method to reduce labeling costs. However, it does not consider fitness among more species. Methods of selecting the most suitable source domain dataset based on the fruit datasets of the target domain remain to be investigated. Moreover, current automatic labeling technology still requires manual labeling of the source domain dataset and cannot completely eliminate manual processes. Therefore, an improved EasyDAM_V3 model was proposed in this study as an automatic labeling method for additional classes of fruit. This study proposes both an optimal source domain establishment method based on a multidimensional spatial feature model to select the most suitable source domain, and a high-volume dataset construction method based on transparent background fruit image translation by constructing a knowledge graph of orchard scene hierarchy component synthesis rules. The EasyDAM_V3 model can automatically obtain fruit label information from the dataset, thereby eliminating manual labeling. To test the proposed method, pear was used as the selected optimal source domain, followed by orange, apple, and tomato as the target domain datasets. The results showed that the average precision of annotation reached 90.94%, 89.78%, and 90.84% for the target datasets, respectively. The EasyDAM_V3 model can obtain the optimal source domain in automatic labeling tasks, thus eliminating the manual labeling process and reducing associated costs and labor.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Plant Phenomics
Multiple-
CiteScore
8.60
自引率
9.20%
发文量
26
审稿时长
14 weeks
期刊介绍:
Plant Phenomics is an Open Access journal published in affiliation with the State Key Laboratory of Crop Genetics & Germplasm Enhancement, Nanjing Agricultural University (NAU) and published by the American Association for the Advancement of Science (AAAS). Like all partners participating in the Science Partner Journal program, Plant Phenomics is editorially independent from the Science family of journals.
The mission of Plant Phenomics is to publish novel research that will advance all aspects of plant phenotyping from the cell to the plant population levels using innovative combinations of sensor systems and data analytics. Plant Phenomics aims also to connect phenomics to other science domains, such as genomics, genetics, physiology, molecular biology, bioinformatics, statistics, mathematics, and computer sciences. Plant Phenomics should thus contribute to advance plant sciences and agriculture/forestry/horticulture by addressing key scientific challenges in the area of plant phenomics.
The scope of the journal covers the latest technologies in plant phenotyping for data acquisition, data management, data interpretation, modeling, and their practical applications for crop cultivation, plant breeding, forestry, horticulture, ecology, and other plant-related domains.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


