减少黑曲霉系列中可接受的物种数量。
摘要
黑曲霉(Aspergillus)系列包含生物技术和医学上的重要菌种。它们可以产生有害的霉菌毒素,这与这些菌种经常出现在食品和室内环境中有关。该系列的分类法经历了多次重新整理,目前该系列共有 14 个物种,其中大多数被认为是隐性物种。然而,即使使用 DNA 测序法或 MALDI-TOF 质谱法,许多分离物的种级鉴定仍存在问题或无法鉴定,这表明在物种界限的定义上可能存在问题,或存在未描述的物种多样性。为了重新研究物种界限,我们收集了尼格里系列中 276 个菌株的三个系统发育标记(benA、CaM 和 RPB2)的 DNA 序列,并生成了 18 个新的全基因组序列。利用三基因数据集,我们采用了基于多物种凝聚模型的系统发生方法,包括四种单焦点方法(GMYC、bGMYC、PTP 和 bPTP)和一种多焦点方法(STACEY)。在总共 15 种方法及其各种设置中,有 11 种只支持识别与三个主要系统发生系相对应的三个物种:A. niger、A. tubingensis 和 A. brasiliensis。同样,GCPSR 方法(谱系一致系统发育物种识别)和 DELINEATE 软件的分析也支持识别这三个物种。我们还发现,基于 benA、CaM 和 RPB2 的系统发生是次优的,与使用 5 752 个单拷贝直向同源蛋白构建的系统发生存在显著差异;因此,划分方法的结果可能会受到比通常水平更高的不确定性的影响。为了克服这一问题,我们从这些基因组中随机选择了 200 个基因,进行了 10 次独立的 STACEY 分析,每次分析 20 个基因。所有分析结果都支持在 A. niger 和 A. brasiliensis 支系中只识别一个物种,而在 A. tubingensis 支系中则不一致地划分出一至四个物种。在考虑了所有这些结果及其实际影响后,我们建议修订后的 Nigri 系列包括 6 个种:A. brasiliensis、A. eucalypticola、A. luchuensis(与 A. piperis 同源)、A. niger(与 A. vinaceus 和 A. welwitschiae 同源)、A. tubingensis(与 A. chiangmaiensis、A. costaricensis、A. neoniger 和 A. pseudopiperis 同源)和 A. vadensis。我们还发现,重新定义的 A. niger 和 A. tubingensis 的种内遗传变异性与其他天青霉菌中常见的遗传变异性并无差异。我们在研究中补充了一份已被接受的种、异名和未解决名称的清单,其中一些名称可能会威胁到当前分类法的稳定性。引用:Bian C, Kusuya Y, Sklenář F, D'hooge E, Yaguchi T, Ban S, Visagie CM, Houbraken J, Takahashi H, Hubka V (2022)。减少黑曲霉系列中可接受菌种的数量。Doi: 10.3114/sim.2022.102.03.


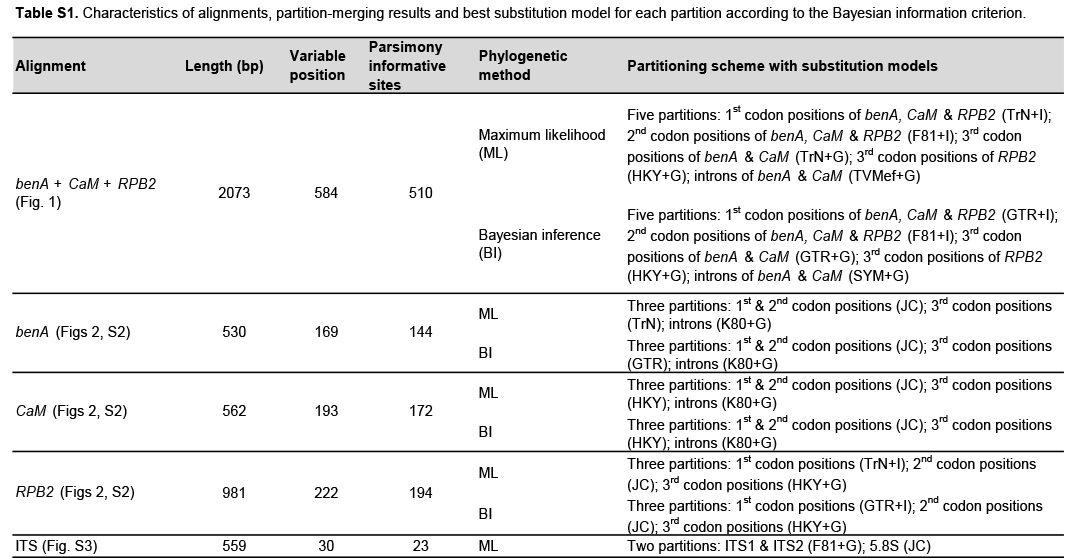
The Aspergillus series Nigri contains biotechnologically and medically important species. They can produce hazardous mycotoxins, which is relevant due to the frequent occurrence of these species on foodstuffs and in the indoor environment. The taxonomy of the series has undergone numerous rearrangements, and currently, there are 14 species accepted in the series, most of which are considered cryptic. Species-level identifications are, however, problematic or impossible for many isolates even when using DNA sequencing or MALDI-TOF mass spectrometry, indicating a possible problem in the definition of species limits or the presence of undescribed species diversity. To re-examine the species boundaries, we collected DNA sequences from three phylogenetic markers (benA, CaM and RPB2) for 276 strains from series Nigri and generated 18 new whole-genome sequences. With the three-gene dataset, we employed phylogenetic methods based on the multispecies coalescence model, including four single-locus methods (GMYC, bGMYC, PTP and bPTP) and one multilocus method (STACEY). From a total of 15 methods and their various settings, 11 supported the recognition of only three species corresponding to the three main phylogenetic lineages: A. niger, A. tubingensis and A. brasiliensis. Similarly, recognition of these three species was supported by the GCPSR approach (Genealogical Concordance Phylogenetic Species Recognition) and analysis in DELINEATE software. We also showed that the phylogeny based on benA, CaM and RPB2 is suboptimal and displays significant differences from a phylogeny constructed using 5 752 single-copy orthologous proteins; therefore, the results of the delimitation methods may be subject to a higher than usual level of uncertainty. To overcome this, we randomly selected 200 genes from these genomes and performed ten independent STACEY analyses, each with 20 genes. All analyses supported the recognition of only one species in the A. niger and A. brasiliensis lineages, while one to four species were inconsistently delimited in the A. tubingensis lineage. After considering all of these results and their practical implications, we propose that the revised series Nigri includes six species: A. brasiliensis, A. eucalypticola, A. luchuensis (syn. A. piperis), A. niger (syn. A. vinaceus and A. welwitschiae), A. tubingensis (syn. A. chiangmaiensis, A. costaricensis, A. neoniger and A. pseudopiperis) and A. vadensis. We also showed that the intraspecific genetic variability in the redefined A. niger and A. tubingensis does not deviate from that commonly found in other aspergilli. We supplemented the study with a list of accepted species, synonyms and unresolved names, some of which may threaten the stability of the current taxonomy. Citation: Bian C, Kusuya Y, Sklenář F, D'hooge E, Yaguchi T, Ban S, Visagie CM, Houbraken J, Takahashi H, Hubka V (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology 102: 95-132. doi: 10.3114/sim.2022.102.03.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


