基于无标记策略的LC-MS/MS†分析诱导多能干细胞生成的大骨节病软骨细胞n -糖基化蛋白
IF 3
4区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
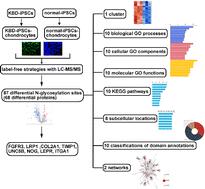
我们的目的是比较大骨节病(KBD)软骨细胞和诱导多能干细胞(iPSCs)衍生的正常软骨细胞中的n -糖基化蛋白。KBD和正常iPSCs分别从人KBD和正常真皮成纤维细胞中重编程。随后,分别从KBD和正常ipsc中分化出软骨细胞。采用免疫荧光法检测诱导多能干细胞和软骨细胞的蛋白标记物。采用LC-MS/MS无标记策略筛选差异n -糖基化蛋白。利用生物信息学分析来解释不同n -糖基化蛋白的功能。免疫荧光染色显示,KBD-iPSCs和正常- ipscs均强烈表达多能性标志物OCT4和NANOG。同时,软骨细胞标志物胶原II和SOX9在kbd - ipsc -软骨细胞和正常- ipsc -软骨细胞中存在。我们得到了87个不同的n -糖基化位点,对应68个不同的蛋白质,并将它们构建成1个簇。我们获得了I型胶原三聚体和其他9个生物过程;多糖结合等9种分子功能;RNA聚合酶II和氧化石墨烯的其他9种细胞成分对转录的调控;Pl3K-Akt信号通路和其他9条KEGG信号通路;过氧化物酶体和其他7个亚细胞位置;整合素α链、c端细胞质区、保守位点等9种结构域注释,2个网络。FGFR3和LRP1在kbd - ipsc -软骨细胞中表达水平较高,而COL2A1、TIMP1、UNC5B、NOG、LEPR和ITGA1在kbd - ipsc -软骨细胞中表达水平下调。这些n -糖基化蛋白的差异表达可能导致大骨节软骨细胞功能异常。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Analysis of N-glycosylation protein of Kashin–Beck disease chondrocytes derived from induced pluripotent stem cells based on label-free strategies with LC-MS/MS†
We aimed to compare N-glycosylation proteins in Kashin-Beck disease (KBD) chondrocytes and normal chondrocytes derived from induced pluripotent stem cells (iPSCs). KBD and normal iPSCs were reprogrammed from human KBD and normal dermal fibroblasts, respectively. Subsequently, chondrocytes were differentiated from KBD and normal iPSCs separately. Immunofluorescence was utilized to assay the protein markers of iPSCs and chondrocytes. Differential N-glycosylation proteins were screened using label-free strategies with LC-MS/MS. Bioinformatics analyses were utilized to interpret the functions of differential N-glycosylation proteins. Immunofluorescence staining revealed that both KBD-iPSCs and normal-iPSCs strongly expressed pluripotency markers OCT4 and NANOG. Meanwhile, chondrocyte markers collagen II and SOX9 are presented in KBD-iPSC-chondrocytes and normal-iPSC-chondrocytes. We obtained 87 differential N-glycosylation sites which corresponded to 68 differential proteins, which were constructed into 1 cluster. We obtained collagen type I trimer and 9 other biological processes; polysaccharide binding and 9 other molecular functions; regulation of transcription by RNA polymerase II and 9 other cellular components from GO; the Pl3K-Akt signaling pathway and 9 other KEGG pathways; peroxisome and 7 other subcellular locations; and integrin alpha chain, C-terminal cytoplasmic region, conserved site and 9 other classifications of domain annotations, and 2 networks. FGFR3 and LRP1 are expressed at higher levels in KBD-iPSC-chondrocytes, while the expressions of COL2A1, TIMP1, UNC5B, NOG, LEPR, and ITGA1 were down-regulated in KBD-iPSC-chondrocytes. The differential expressions of these N-glycosylation proteins may lead to the abnormal function of KBD chondrocytes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular omics
Biochemistry, Genetics and Molecular Biology-Biochemistry
CiteScore
5.40
自引率
3.40%
发文量
91
期刊介绍:
Molecular Omics publishes high-quality research from across the -omics sciences.
Topics include, but are not limited to:
-omics studies to gain mechanistic insight into biological processes – for example, determining the mode of action of a drug or the basis of a particular phenotype, such as drought tolerance
-omics studies for clinical applications with validation, such as finding biomarkers for diagnostics or potential new drug targets
-omics studies looking at the sub-cellular make-up of cells – for example, the subcellular localisation of certain proteins or post-translational modifications or new imaging techniques
-studies presenting new methods and tools to support omics studies, including new spectroscopic/chromatographic techniques, chip-based/array technologies and new classification/data analysis techniques. New methods should be proven and demonstrate an advance in the field.
Molecular Omics only accepts articles of high importance and interest that provide significant new insight into important chemical or biological problems. This could be fundamental research that significantly increases understanding or research that demonstrates clear functional benefits.
Papers reporting new results that could be routinely predicted, do not show a significant improvement over known research, or are of interest only to the specialist in the area are not suitable for publication in Molecular Omics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


