Mallard和Muscovy鸭微卫星标记的全基因组计算机表征、验证和跨物种可转移性。
IF 3.5
Q3 Biochemistry, Genetics and Molecular Biology
Journal of Genetic Engineering and Biotechnology
Pub Date : 2023-12-01
DOI:10.1186/s43141-023-00556-z
引用次数: 0
摘要
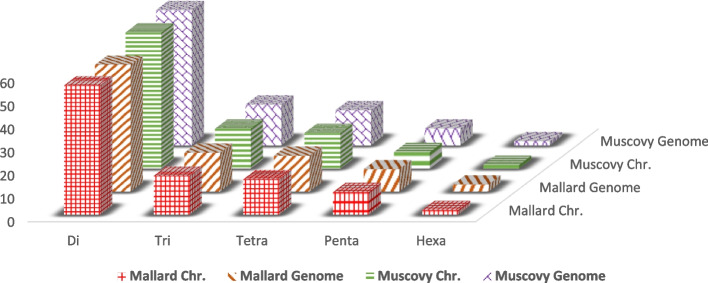
背景:微卫星是包括鸭子在内的家畜的重要标记。微型卫星的开发既昂贵又耗费人力。与此同时,微型卫星的计算机采矿方法成为一种可行的替代方法。因此,本研究旨在比较番鸭和Mallard鸭的全基因组和染色体微卫星挖掘方法,并测试它们之间标记的可转移性。GMATA软件用于计算机研究,并使用26个引物进行验证。结果:Mallards和Muscovies的染色体微卫星总数分别为250053和226417个位点,而全基因组微卫星总数为260059和238462个位点。使用这两种方法,不同图案的频率具有相似的模式。二核苷酸基序占主导地位(> 50%)。基因组扩增显示,Mallards和Muscovies的平均等位基因数分别为5.08和4.96。一个基因座在Mallards中是专态的,两个在Muscovies中是单态的。番鸭的平均预期杂合度高于Mallards(0.45对0.43),两个引物组之间没有显著差异,这表明不同引物的跨物种扩增是有用的。结论:本研究首次为鸭子开发了全基因组SSR图谱,结果可以证明,与全基因组方法相比,染色体挖掘不会产生不同的结果。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Genome-wide in silico characterization, validation, and cross-species transferability of microsatellite markers in Mallard and Muscovy ducks
Background
Microsatellites are important markers for livestock including ducks. The development of microsatellites is expensive and labor-intensive. Meanwhile, the in silico approach for mining for microsatellites became a practicable alternative. Therefore, the current study aimed at comparing whole-genome and chromosome-wise microsatellite mining approaches in Muscovy and Mallard ducks and testing the transferability of markers between them. The GMATA software was used for the in silico study, and validation was performed using 26 primers.
Results
The total number of the detected microsatellites using chromosome-wise was 250,053 and 226,417 loci compared to 260,059 and 238,462 loci using whole genome in Mallards and Muscovies. The frequencies of different motifs had similar patterns using the two approaches. Dinucleotide motifs were predominant (> 50%) in both Mallards and Muscovies. The amplification of the genomes revealed an average number of alleles of 5.08 and 4.96 in Mallards and Muscovies. One locus was monographic in Mallards, and two were monomorphic in Muscovies. The average expected heterozygosity was higher in Muscovy than in Mallards (0.45 vs. 0.43) with no significant difference between the two primer sets, which indicated the usefulness of cross-species amplification of different primers.
Conclusion
The current study developed a whole-genome SSR panel for ducks for the first time, and the results could prove that using chromosome-wise mining did not generate different results compared to the whole-genome approach.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Genetic Engineering and Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
5.70
自引率
5.70%
发文量
159
审稿时长
16 weeks
期刊介绍:
Journal of genetic engineering and biotechnology is devoted to rapid publication of full-length research papers that leads to significant contribution in advancing knowledge in genetic engineering and biotechnology and provide novel perspectives in this research area. JGEB includes all major themes related to genetic engineering and recombinant DNA. The area of interest of JGEB includes but not restricted to: •Plant genetics •Animal genetics •Bacterial enzymes •Agricultural Biotechnology, •Biochemistry, •Biophysics, •Bioinformatics, •Environmental Biotechnology, •Industrial Biotechnology, •Microbial biotechnology, •Medical Biotechnology, •Bioenergy, Biosafety, •Biosecurity, •Bioethics, •GMOS, •Genomic, •Proteomic JGEB accepts
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


