使用Cas12a/3D DNAzyme比色纸传感器检测细菌病原体抗生素耐药基因
IF 6.3
3区 综合性期刊
Q1 Multidisciplinary
引用次数: 0
摘要
在细菌性病原体中快速检测耐抗生素基因对于应对全球卫生危机至关重要。在此,我们报道了一种基于CRISPR/ cas12的比色纸传感器,其中Cas12a的反式切割活性通过滚动圈复制后扩增,从而产生3D DNAzyme。3D DNAzyme牢固地粘附在纸张表面,创造了一种具有高生物活性的纸张传感器,其中包含高密度的功能性DNAzyme。该方法可快速检测耐药基因NDM-1,灵敏度高。当NDM-1基因缺失时,3D DNAzyme催化比色反应,产生蓝色信号,而当NDM-1存在时,Cas12a侧支裂解活性被激活,导致圆环模板被切割,从而阻止3D DNAzyme的产生,不产生比色信号。该传感器能够快速、低成本地检测各种病原微生物携带的耐药基因,具有飞摩尔级的灵敏度和肉眼可见的结果。整个分析需要不到90分钟的分析时间。由于CRISPR探针的高度可编程设计,该平台具有对新的全球流行病快速反应的巨大潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
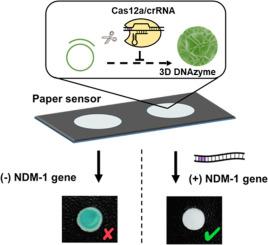
Detection of antibiotic-resistance genes in bacterial pathogens using a Cas12a/3D DNAzyme colorimetric paper sensor
The rapid detection of antibiotic-resistant genes in bacterial pathogens is critical in combating global health crises. Herein, we report a CRISPR/Cas12a-based colorimetric paper sensor, where the trans-cleavage activity of Cas12a was post-amplified by rolling circle replication, resulting in the generation of a 3D DNAzyme. The 3D DNAzyme adhered strongly to the paper surface, creating a highly bioactive paper sensor containing high densities of functional DNAzymes. This assay was effective for the rapid detection of the antibiotic-resistant gene, NDM-1, with high sensitivity. In the absence of the NDM-1 gene, the 3D DNAzyme catalyzed a colorimetric reaction, resulting in a blue-colored signal while in the presence of NDM-1, collateral cleavage activity of Cas12a was activated, leading to cleavage of the circle template, thus preventing the generation of the 3D DNAzyme and producing no colorimetric signal. This paper sensor provides rapid and low-cost detection of antibiotic-resistant genes carried by various pathogenic microorganisms with femtomolar-level sensitivity and results that are visible to the naked eye. The entire analysis requires less than 90 minutes of assay time. Due to the highly programmable design of the CRISPR probe, the platform has significant potential for quick responses to new global epidemics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Fundamental Research
Multidisciplinary-Multidisciplinary
CiteScore
4.00
自引率
1.60%
发文量
294
审稿时长
79 days
期刊介绍:
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


