使用健康的单细胞参照物精确鉴定疾病中改变的细胞状态。
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
联合分析来自患病组织和健康参考的单细胞基因组学数据可以揭示细胞状态的改变。我们调查了来自健康个体的综合数据集(细胞图谱)是否适合作为疾病状态识别的参考,以及是否需要匹配的对照样本来最大限度地减少错误发现。我们证明,使用参考图谱进行潜在空间学习,然后对匹配的对照进行差异分析,可以改进对疾病相关细胞的识别,尤其是对多种受干扰细胞类型的识别。此外,当图谱可用时,减少对照样本数量不会增加错误发现率。通过联合分析新冠肺炎队列和血细胞图谱的数据,我们改进了与不同临床严重程度相关的感染相关细胞状态的检测。同样,我们使用健康肺图谱研究了肺纤维化的疾病状态,表征了两种不同的异常基础状态。我们的分析为设计疾病队列研究和优化细胞图谱的使用提供了指导。本文章由计算机程序翻译,如有差异,请以英文原文为准。
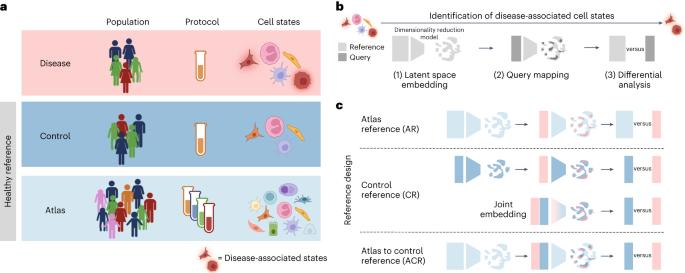
Precise identification of cell states altered in disease using healthy single-cell references
Joint analysis of single-cell genomics data from diseased tissues and a healthy reference can reveal altered cell states. We investigate whether integrated collections of data from healthy individuals (cell atlases) are suitable references for disease-state identification and whether matched control samples are needed to minimize false discoveries. We demonstrate that using a reference atlas for latent space learning followed by differential analysis against matched controls leads to improved identification of disease-associated cells, especially with multiple perturbed cell types. Additionally, when an atlas is available, reducing control sample numbers does not increase false discovery rates. Jointly analyzing data from a COVID-19 cohort and a blood cell atlas, we improve detection of infection-related cell states linked to distinct clinical severities. Similarly, we studied disease states in pulmonary fibrosis using a healthy lung atlas, characterizing two distinct aberrant basal states. Our analysis provides guidelines for designing disease cohort studies and optimizing cell atlas use. In single-cell studies, combining healthy reference atlases and designed control datasets allows more precise identification of disease-associated cell states.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


