Levenstein距离、序列比对与生物数据库检索
IF 2.2
3区 计算机科学
Q3 COMPUTER SCIENCE, INFORMATION SYSTEMS
引用次数: 39
摘要
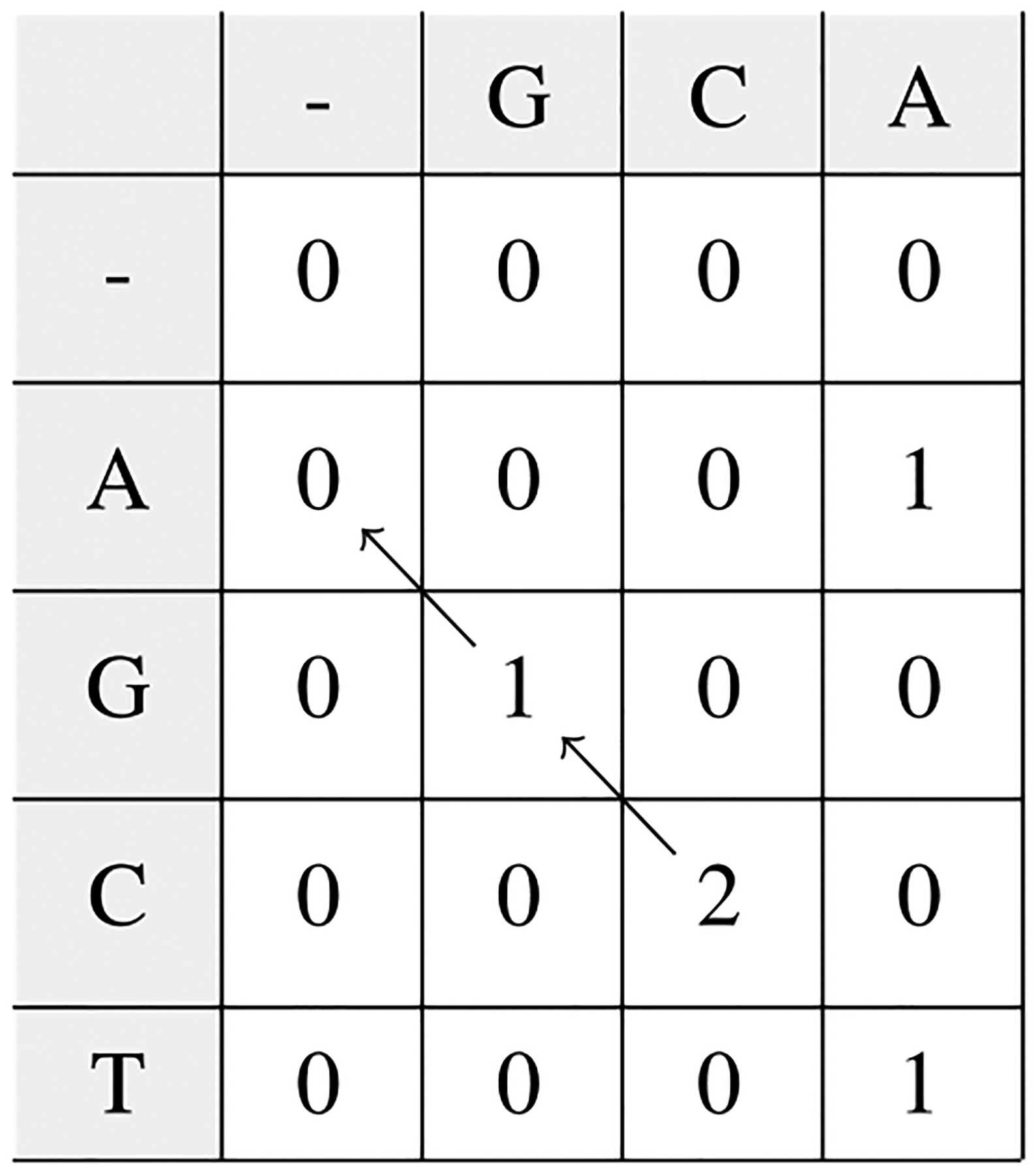
Levenstein编辑距离在过去和现在的序列比对和生物数据库相似性搜索中都发挥着核心作用。我们从计算Levenstein距离和序列比对的动态规划算法的历史开始回顾。接下来,我们描述了这些算法是如何导致启发式算法在生物信息学中最广泛使用的软件BLAST中使用的,BLAST是一个搜索DNA和蛋白质数据库以寻找进化相关相似性的程序。最近,现代基因组测序的出现及其产生的大量数据导致了局部比对问题的回归。最后,我们总结了Levenstein距离作为度量的数学公式如何使生物环境中的相似性搜索有可能得到额外的优化。这些现代优化是围绕生物数据库的低度量熵和分数维构建的,实现了生物相似性搜索的数量级加速。本文章由计算机程序翻译,如有差异,请以英文原文为准。
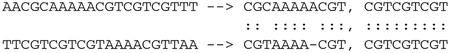
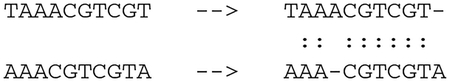
Levenshtein Distance, Sequence Comparison and Biological Database Search
Levenshtein edit distance has played a central role-both past and present-in sequence alignment in particular and biological database similarity search in general. We start our review with a history of dynamic programming algorithms for computing Levenshtein distance and sequence alignments. Following, we describe how those algorithms led to heuristics employed in the most widely used software in bioinformatics, BLAST, a program to search DNA and protein databases for evolutionarily relevant similarities. More recently, the advent of modern genomic sequencing and the volume of data it generates has resulted in a return to the problem of local alignment. We conclude with how the mathematical formulation of Levenshtein distance as a metric made possible additional optimizations to similarity search in biological contexts. These modern optimizations are built around the low metric entropy and fractional dimensionality of biological databases, enabling orders of magnitude acceleration of biological similarity search.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

IEEE Transactions on Information Theory
工程技术-工程:电子与电气
CiteScore
5.70
自引率
20.00%
发文量
514
审稿时长
12 months
期刊介绍:
The IEEE Transactions on Information Theory is a journal that publishes theoretical and experimental papers concerned with the transmission, processing, and utilization of information. The boundaries of acceptable subject matter are intentionally not sharply delimited. Rather, it is hoped that as the focus of research activity changes, a flexible policy will permit this Transactions to follow suit. Current appropriate topics are best reflected by recent Tables of Contents; they are summarized in the titles of editorial areas that appear on the inside front cover.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


