一种来自 SEPALLATA3 的 circRNA 通过 R 环的形成调节其同源 mRNA 的剪接
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 404
摘要
环状 RNA(circRNA)是一类丰富多样的超稳定、非规范 RNA,通过一种称为反向剪接的替代剪接(AS)形式产生。这些单链、共价闭合的 circRNA 分子已在所有真核生物界中被发现1,但它们的功能仍然难以捉摸。在这里,我们报告了 circRNA 可作为拟南芥中功能性外显子断裂 AS 变异的真正生物标志物,包括在同源的 MADS-box 转录因子家族中。此外,我们还证明了源自SEPALLATA3(SEP3)基因第6外显子的circRNA会增加同源的外显子缺失AS变体(缺乏第6外显子的SEP3.3)的丰度,进而驱动花卉同源表型。为了证明其基本机制,我们发现 SEP3 第 6 外显子 circRNA 能与其同源的 DNA 基因座紧密结合,形成 RNA:DNA 混合体或 R 环,而线性 RNA 等价物与 DNA 的结合力要弱得多。R-loop 的形成会导致转录暂停,这已被证明与剪接因子的招募和 AS2-4 相吻合。本报告提出了一种新的机理观点,即至少有一部分环状 RNA 可能有助于提高其同源外显子跳断信使 RNA 的剪接效率,并首次提供了由环状 RNA 操作介导的生物体水平表型的证据。虽然在所有真核生物界都发现了环状 RNA(circRNA),但它们的功能仍然难以捉摸。现在,一项研究表明,circRNA 可促进其同源 mRNA 的替代剪接,从而驱动同源表型。本文章由计算机程序翻译,如有差异,请以英文原文为准。
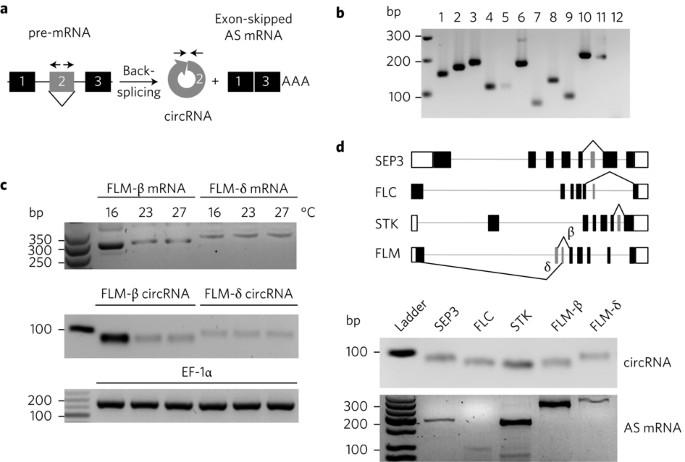
A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation
Circular RNAs (circRNAs) are a diverse and abundant class of hyper-stable, non-canonical RNAs that arise through a form of alternative splicing (AS) called back-splicing. These single-stranded, covalently-closed circRNA molecules have been identified in all eukaryotic kingdoms of life1, yet their functions have remained elusive. Here, we report that circRNAs can be used as bona fide biomarkers of functional, exon-skipped AS variants in Arabidopsis, including in the homeotic MADS-box transcription factor family. Furthermore, we demonstrate that circRNAs derived from exon 6 of the SEPALLATA3 (SEP3) gene increase abundance of the cognate exon-skipped AS variant (SEP3.3 which lacks exon 6), in turn driving floral homeotic phenotypes. Toward demonstrating the underlying mechanism, we show that the SEP3 exon 6 circRNA can bind strongly to its cognate DNA locus, forming an RNA:DNA hybrid, or R-loop, whereas the linear RNA equivalent bound significantly more weakly to DNA. R-loop formation results in transcriptional pausing, which has been shown to coincide with splicing factor recruitment and AS2–4. This report presents a novel mechanistic insight for how at least a subset of circRNAs probably contribute to increased splicing efficiency of their cognate exon-skipped messenger RNA and provides the first evidence of an organismal-level phenotype mediated by circRNA manipulation. While circular RNAs (circRNAs) have been identified in all eukaryotic kingdoms of life, their functions have remained elusive. Now, a study shows that circRNAs can promote alternative splicing of their cognate mRNA, thus driving homeotic phenotypes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


