用Maldi质谱法测定大肠杆菌异亮氨酸- trna合成酶的一级结构和活性位点。
摘要
利用4种内源性蛋白酶(trypsin、LysC、AspN和GluC),采用MALDI质谱法,建立了大肠杆菌异亮氨酸- trna合成酶(IleRS)的正确氨基酸序列。随后,利用底物的活性类似物进行亲和标记,绘制了IleRS的活性位点。对于ATP结合位点,亲和标记试剂为吡哆醛5′-二磷酸-5′-腺苷(ADP-PL),而高碘酸氧化tRNA(Ile), tRNA(Ile)的2′,3′-双醛衍生物(Ile)用于标记tRNA在合成酶上的3′端结合位点。任何一种试剂与IleRS孵育都导致tRNA(Ile)氨基酰化和异亮氨酸依赖的同位素ATP-PPi交换活性迅速丧失。用ADP-PL或tRNA(Ile)ox标记的IleRS的化学计量量对应于每mol酶掺入1 mol试剂。总的来说,tRNA的氧化3'端(Ile)和ATP类似物ADP-PL的吡哆醛部分与一致序列(601)KMSKS(605)的赖氨酸残基601和604发生反应。通过与l -异亮氨酸的溴甲基酮衍生物(IBMK)或非同源氨基酸缬氨酸(VBMK)、苯丙氨酸(FBMK)和去甲亮氨酸(NleBMK)的合成酶的定性比较标记,鉴定了大肠杆菌上l -异亮氨酸或非同源氨基酸的结合位点。用IBMK标记酶导致完全丧失异亮氨酸依赖的同位素[(32)P]PPi-ATP交换活性。VBMK、NleBMK和FBMK也能消除IleRS的活性,FBMK在灭活合成酶方面效率较低。MALDI质谱分析表明,底物类似物IBMK在大肠杆菌上的靶残基为半胱氨酸-462和-718,而VBMK、NleBMK和FBMK标记的是His-394、His-478和Cys-718。此外,化学性质与IBMK相似的VBMK和NleBMK被发现与Cys-462共价结合,VBMK特异性地附着在合成酶的His-332(或His-337)上。底物类似物标记的氨基酸残基主要分布在大肠杆菌一级结构的三个区域:片段[325-394]、[451-479]和[591-604]。在来自T. thermophilus和S. aureus的IleRS的三维结构中,[325-394]片段是编辑结构域的一部分,而片段[451-479]和[591-604]分别代表异亮氨酸结合结构域和二核苷酸(或Rossmann)折叠结构域位于催化核心。大肠杆菌的His-332位于合成酶的编辑活性位点,在所有可用的大肠杆菌序列中严格保守。据推测,大肠杆菌的His-332直接参与水解,或在水解位点帮助苏氨酸的羟基去质子化。


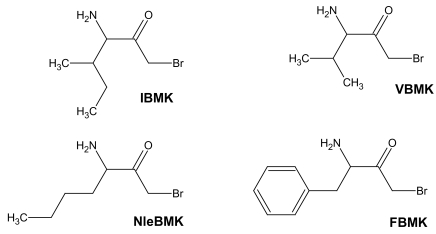
The correct amino acid sequence of E. coli isoleucyl-tRNA synthetase (IleRS) was established by means of peptide mapping by MALDI mass spectrometry, using a set of four endoproteases (trypsin, LysC, AspN and GluC). Thereafter, the active site of IleRS was mapped by affinity labeling with reactive analogs of the substrates. For the ATP binding site, the affinity labeling reagent was pyridoxal 5'-diphospho-5'-adenosine (ADP-PL), whereas periodate-oxidized tRNA(Ile), the 2',3'-dialdehyde derivative of tRNA(Ile) was used to label the binding site for the 3'-end of tRNA on the synthetase. Incubation of either reagent with IleRS resulted in a rapid loss of both the tRNA(Ile) aminoacylation and isoleucinedependent isotopic ATP-PPi exchange activities. The stoichiometries of IleRS labeling by ADP-PL or tRNA(Ile)ox corresponded to 1 mol of reagent incorporated per mol of enzyme. Altogether, the oxidized 3'-end of tRNA(Ile) and the pyridoxal moiety of the ATP analog ADP-PL react with the lysyl residues 601 and 604 of the consensus sequence (601)KMSKS(605). Identification of the binding site for L-isoleucine or for non cognate amino acids on E. coli IleRS was achieved by qualitative comparative labeling of the synthetase with bromomethyl ketone derivatives of L-isoleucine (IBMK) or of the non-cognate amino acids valine (VBMK), phenylalanine (FBMK) and norleucine (NleBMK). Labeling of the enzyme with IBMK resulted in a complete loss of isoleucine-dependent isotopic [(32)P]PPi-ATP exchange activity. VBMK, NleBMK and FBMK were also capable of abolishing the activity of IleRS, FBMK being the less efficient in inactivating the synthetase. Analysis by MALDI mass spectrometry designated cysteines-462 and -718 as the target residues of the substrate analog IBMK on E. coli IleRS, whereas VBMK, NleBMK and FBMK labeled in common His-394, His-478 and Cys-718. In addition, VBMK and NleBMK, which are chemically similar to IBMK, were found covalently bound to Cys-462, and VBMK was specifically attached to His-332 (or His-337) of the synthetase. The amino acid residues labeled by the substrate analogs are mainly distributed between three regions in the primary structure of E. coli IleRS: these are segments [325-394], [451-479] and [591-604]. In the 3-D structures of IleRS from T. thermophilus and S. aureus, the [325-394] stretch is part of the editing domain, while fragments [451-479] and [591-604] representing the isoleucine binding domain and the dinucleotide (or Rossmann) fold domain, respectively, are located in the catalytic core. His-332 of E. coli IleRS, that is strictly conserved among all the available IleRS sequences is located in the editing active site of the synthetase. It is proposed that His-332 of E. coli IleRS participates directly in hydrolysis, or helps to deprotonate the hydroxyl group of threonine at the hydrolytic site.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


